
Parainfluenza virus 5 (strain W3) (PIV5) (Simian virus 5)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Orthorubulavirus; Mammalian orthorubulavirus 5
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
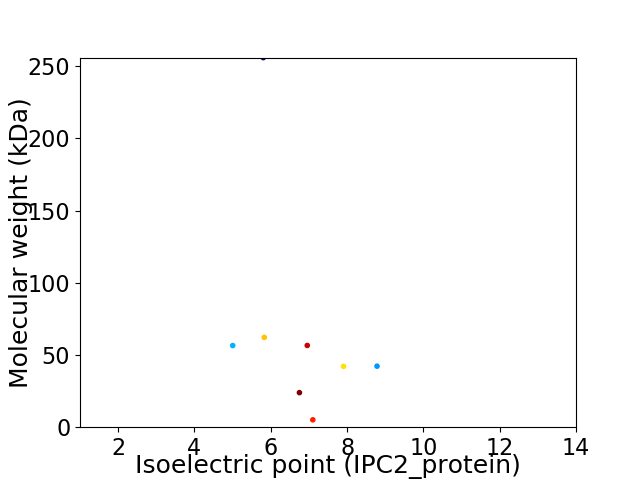
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q88435|NCAP_PIV5 Nucleoprotein OS=Parainfluenza virus 5 (strain W3) OX=11208 GN=NP PE=1 SV=1
MM1 pKa = 7.56SSVLKK6 pKa = 10.47AYY8 pKa = 9.85EE9 pKa = 4.19RR10 pKa = 11.84FTLTQEE16 pKa = 4.31LQDD19 pKa = 3.52QSEE22 pKa = 4.75EE23 pKa = 4.22GTIPPTTLKK32 pKa = 10.47PVIRR36 pKa = 11.84VFILTSNNPEE46 pKa = 3.83LRR48 pKa = 11.84SRR50 pKa = 11.84LLLFCLRR57 pKa = 11.84IVLSNGARR65 pKa = 11.84DD66 pKa = 3.57SHH68 pKa = 7.35RR69 pKa = 11.84FGALLTMFSLPSATMLNHH87 pKa = 6.25VKK89 pKa = 10.41LADD92 pKa = 3.6QSPEE96 pKa = 3.53ADD98 pKa = 3.19IEE100 pKa = 4.16RR101 pKa = 11.84VEE103 pKa = 3.83IDD105 pKa = 3.27GFEE108 pKa = 4.04EE109 pKa = 5.24GSFRR113 pKa = 11.84LIPNARR119 pKa = 11.84SGMSRR124 pKa = 11.84GEE126 pKa = 3.88INAYY130 pKa = 9.8AALAEE135 pKa = 4.73DD136 pKa = 5.71LPDD139 pKa = 4.1TLNHH143 pKa = 6.05ATPFVDD149 pKa = 4.9SEE151 pKa = 4.92VEE153 pKa = 3.95GTAWDD158 pKa = 4.18EE159 pKa = 4.14IEE161 pKa = 4.68TFLDD165 pKa = 3.08MCYY168 pKa = 10.57SVLMQAWIVTCKK180 pKa = 10.52CMTAPDD186 pKa = 3.62QPAASIEE193 pKa = 3.98KK194 pKa = 10.06RR195 pKa = 11.84LQKK198 pKa = 10.3YY199 pKa = 8.17RR200 pKa = 11.84QQGRR204 pKa = 11.84INPRR208 pKa = 11.84YY209 pKa = 9.48LLQPEE214 pKa = 4.17ARR216 pKa = 11.84RR217 pKa = 11.84IIQNVIRR224 pKa = 11.84KK225 pKa = 9.26GMVVRR230 pKa = 11.84HH231 pKa = 5.88FLTFEE236 pKa = 4.09LQLARR241 pKa = 11.84AQSLVSNRR249 pKa = 11.84YY250 pKa = 7.77YY251 pKa = 11.74AMVGDD256 pKa = 3.57VGKK259 pKa = 10.91YY260 pKa = 9.56IEE262 pKa = 4.32NCGMGGFFLTLKK274 pKa = 10.07YY275 pKa = 10.78ALGTRR280 pKa = 11.84WPTLALAAFSGEE292 pKa = 4.03LTKK295 pKa = 10.94LKK297 pKa = 10.92SLMALYY303 pKa = 8.09QTLGEE308 pKa = 4.1QARR311 pKa = 11.84YY312 pKa = 8.72LALLEE317 pKa = 4.94SPHH320 pKa = 8.16LMDD323 pKa = 5.63FAAANYY329 pKa = 7.83PLLYY333 pKa = 9.88SYY335 pKa = 11.71AMGIGYY341 pKa = 9.82VLDD344 pKa = 3.66VNMRR348 pKa = 11.84NYY350 pKa = 10.82AFSRR354 pKa = 11.84SYY356 pKa = 10.23MNKK359 pKa = 9.14TYY361 pKa = 10.16FQLGMEE367 pKa = 4.83TARR370 pKa = 11.84KK371 pKa = 7.78QQGAVDD377 pKa = 3.49MRR379 pKa = 11.84MAEE382 pKa = 4.4DD383 pKa = 3.8LGLTQAEE390 pKa = 4.23RR391 pKa = 11.84TEE393 pKa = 4.24MANTLAKK400 pKa = 9.63LTTANRR406 pKa = 11.84GADD409 pKa = 3.22TRR411 pKa = 11.84GGVNPFSSVTGTTQVPAAATGDD433 pKa = 3.78TLEE436 pKa = 5.22SYY438 pKa = 9.57MAADD442 pKa = 4.53RR443 pKa = 11.84LRR445 pKa = 11.84QRR447 pKa = 11.84YY448 pKa = 9.11ADD450 pKa = 4.45AGTHH454 pKa = 6.39DD455 pKa = 5.64DD456 pKa = 4.11EE457 pKa = 4.72MPPLEE462 pKa = 4.75EE463 pKa = 4.59EE464 pKa = 4.57EE465 pKa = 4.82EE466 pKa = 4.78DD467 pKa = 3.84DD468 pKa = 3.74TSAGPRR474 pKa = 11.84TGPTLEE480 pKa = 4.1QVALDD485 pKa = 3.73IQNAAVGAPIHH496 pKa = 6.48TDD498 pKa = 3.42DD499 pKa = 5.38LNAALGDD506 pKa = 3.89LDD508 pKa = 3.57II509 pKa = 6.39
MM1 pKa = 7.56SSVLKK6 pKa = 10.47AYY8 pKa = 9.85EE9 pKa = 4.19RR10 pKa = 11.84FTLTQEE16 pKa = 4.31LQDD19 pKa = 3.52QSEE22 pKa = 4.75EE23 pKa = 4.22GTIPPTTLKK32 pKa = 10.47PVIRR36 pKa = 11.84VFILTSNNPEE46 pKa = 3.83LRR48 pKa = 11.84SRR50 pKa = 11.84LLLFCLRR57 pKa = 11.84IVLSNGARR65 pKa = 11.84DD66 pKa = 3.57SHH68 pKa = 7.35RR69 pKa = 11.84FGALLTMFSLPSATMLNHH87 pKa = 6.25VKK89 pKa = 10.41LADD92 pKa = 3.6QSPEE96 pKa = 3.53ADD98 pKa = 3.19IEE100 pKa = 4.16RR101 pKa = 11.84VEE103 pKa = 3.83IDD105 pKa = 3.27GFEE108 pKa = 4.04EE109 pKa = 5.24GSFRR113 pKa = 11.84LIPNARR119 pKa = 11.84SGMSRR124 pKa = 11.84GEE126 pKa = 3.88INAYY130 pKa = 9.8AALAEE135 pKa = 4.73DD136 pKa = 5.71LPDD139 pKa = 4.1TLNHH143 pKa = 6.05ATPFVDD149 pKa = 4.9SEE151 pKa = 4.92VEE153 pKa = 3.95GTAWDD158 pKa = 4.18EE159 pKa = 4.14IEE161 pKa = 4.68TFLDD165 pKa = 3.08MCYY168 pKa = 10.57SVLMQAWIVTCKK180 pKa = 10.52CMTAPDD186 pKa = 3.62QPAASIEE193 pKa = 3.98KK194 pKa = 10.06RR195 pKa = 11.84LQKK198 pKa = 10.3YY199 pKa = 8.17RR200 pKa = 11.84QQGRR204 pKa = 11.84INPRR208 pKa = 11.84YY209 pKa = 9.48LLQPEE214 pKa = 4.17ARR216 pKa = 11.84RR217 pKa = 11.84IIQNVIRR224 pKa = 11.84KK225 pKa = 9.26GMVVRR230 pKa = 11.84HH231 pKa = 5.88FLTFEE236 pKa = 4.09LQLARR241 pKa = 11.84AQSLVSNRR249 pKa = 11.84YY250 pKa = 7.77YY251 pKa = 11.74AMVGDD256 pKa = 3.57VGKK259 pKa = 10.91YY260 pKa = 9.56IEE262 pKa = 4.32NCGMGGFFLTLKK274 pKa = 10.07YY275 pKa = 10.78ALGTRR280 pKa = 11.84WPTLALAAFSGEE292 pKa = 4.03LTKK295 pKa = 10.94LKK297 pKa = 10.92SLMALYY303 pKa = 8.09QTLGEE308 pKa = 4.1QARR311 pKa = 11.84YY312 pKa = 8.72LALLEE317 pKa = 4.94SPHH320 pKa = 8.16LMDD323 pKa = 5.63FAAANYY329 pKa = 7.83PLLYY333 pKa = 9.88SYY335 pKa = 11.71AMGIGYY341 pKa = 9.82VLDD344 pKa = 3.66VNMRR348 pKa = 11.84NYY350 pKa = 10.82AFSRR354 pKa = 11.84SYY356 pKa = 10.23MNKK359 pKa = 9.14TYY361 pKa = 10.16FQLGMEE367 pKa = 4.83TARR370 pKa = 11.84KK371 pKa = 7.78QQGAVDD377 pKa = 3.49MRR379 pKa = 11.84MAEE382 pKa = 4.4DD383 pKa = 3.8LGLTQAEE390 pKa = 4.23RR391 pKa = 11.84TEE393 pKa = 4.24MANTLAKK400 pKa = 9.63LTTANRR406 pKa = 11.84GADD409 pKa = 3.22TRR411 pKa = 11.84GGVNPFSSVTGTTQVPAAATGDD433 pKa = 3.78TLEE436 pKa = 5.22SYY438 pKa = 9.57MAADD442 pKa = 4.53RR443 pKa = 11.84LRR445 pKa = 11.84QRR447 pKa = 11.84YY448 pKa = 9.11ADD450 pKa = 4.45AGTHH454 pKa = 6.39DD455 pKa = 5.64DD456 pKa = 4.11EE457 pKa = 4.72MPPLEE462 pKa = 4.75EE463 pKa = 4.59EE464 pKa = 4.57EE465 pKa = 4.82EE466 pKa = 4.78DD467 pKa = 3.84DD468 pKa = 3.74TSAGPRR474 pKa = 11.84TGPTLEE480 pKa = 4.1QVALDD485 pKa = 3.73IQNAAVGAPIHH496 pKa = 6.48TDD498 pKa = 3.42DD499 pKa = 5.38LNAALGDD506 pKa = 3.89LDD508 pKa = 3.57II509 pKa = 6.39
Molecular weight: 56.54 kDa
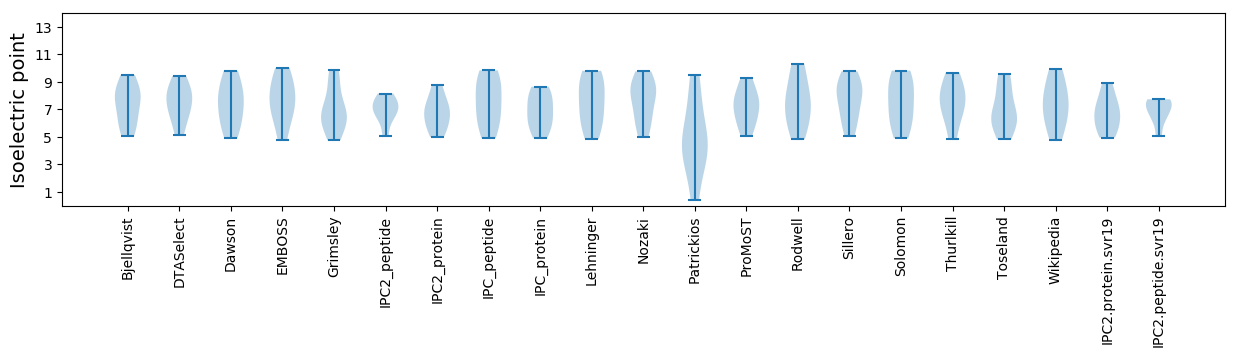
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q88434|L_PIV5 RNA-directed RNA polymerase L OS=Parainfluenza virus 5 (strain W3) OX=11208 GN=L PE=1 SV=1
MM1 pKa = 7.68PSISIPADD9 pKa = 3.13PTNPRR14 pKa = 11.84QSIKK18 pKa = 10.76AFPIVINSDD27 pKa = 3.08GGEE30 pKa = 3.91KK31 pKa = 10.56GRR33 pKa = 11.84LVKK36 pKa = 9.9QLRR39 pKa = 11.84TTYY42 pKa = 11.12LNDD45 pKa = 4.07LDD47 pKa = 3.93THH49 pKa = 6.37EE50 pKa = 4.97PLVTFINTYY59 pKa = 10.24GFIYY63 pKa = 10.18EE64 pKa = 3.99QDD66 pKa = 3.28RR67 pKa = 11.84GNTIVGEE74 pKa = 4.08DD75 pKa = 3.34QLGKK79 pKa = 10.38KK80 pKa = 9.83RR81 pKa = 11.84EE82 pKa = 4.16AVTAAMVTLGCGPNLPSLGNVLGQLRR108 pKa = 11.84EE109 pKa = 4.0FQVTVRR115 pKa = 11.84KK116 pKa = 7.95TSSKK120 pKa = 9.45AEE122 pKa = 3.74EE123 pKa = 4.02MVFEE127 pKa = 4.11IVKK130 pKa = 9.73YY131 pKa = 8.79PRR133 pKa = 11.84IFRR136 pKa = 11.84GHH138 pKa = 5.43TLIQKK143 pKa = 10.21GLVCVSAEE151 pKa = 4.2KK152 pKa = 10.22FVKK155 pKa = 10.57SPGKK159 pKa = 9.4IQSGMDD165 pKa = 3.34YY166 pKa = 11.12LFIPTFLSVTYY177 pKa = 9.89CPAAIKK183 pKa = 9.84FQVPGPMLKK192 pKa = 8.94MRR194 pKa = 11.84SRR196 pKa = 11.84YY197 pKa = 6.47TQSLQLEE204 pKa = 4.25LMIRR208 pKa = 11.84ILCKK212 pKa = 9.79PDD214 pKa = 3.31SPLMKK219 pKa = 10.14VHH221 pKa = 6.4TPDD224 pKa = 3.67KK225 pKa = 10.25EE226 pKa = 4.28GRR228 pKa = 11.84GCLVSVWLHH237 pKa = 4.57VCNIFKK243 pKa = 10.67SGNKK247 pKa = 9.49NGSEE251 pKa = 3.74WQEE254 pKa = 3.32YY255 pKa = 7.21WMRR258 pKa = 11.84KK259 pKa = 7.19CANMQLEE266 pKa = 4.3VSIADD271 pKa = 3.19MWGPTIIIHH280 pKa = 6.41ARR282 pKa = 11.84GHH284 pKa = 5.71IPKK287 pKa = 9.82SAKK290 pKa = 10.39LFFGKK295 pKa = 10.44GGWSCHH301 pKa = 5.65PLHH304 pKa = 6.51EE305 pKa = 4.59VVPSVTKK312 pKa = 8.79TLWSVGCEE320 pKa = 3.51ITKK323 pKa = 10.6AKK325 pKa = 10.62AIIQEE330 pKa = 4.25SSISLLVEE338 pKa = 3.81TTDD341 pKa = 3.97IISPKK346 pKa = 10.19VKK348 pKa = 9.79ISSKK352 pKa = 9.77HH353 pKa = 4.19RR354 pKa = 11.84RR355 pKa = 11.84FVKK358 pKa = 10.78SNWGLFKK365 pKa = 10.08KK366 pKa = 9.01TKK368 pKa = 10.1SLPNLTEE375 pKa = 4.03LEE377 pKa = 4.23
MM1 pKa = 7.68PSISIPADD9 pKa = 3.13PTNPRR14 pKa = 11.84QSIKK18 pKa = 10.76AFPIVINSDD27 pKa = 3.08GGEE30 pKa = 3.91KK31 pKa = 10.56GRR33 pKa = 11.84LVKK36 pKa = 9.9QLRR39 pKa = 11.84TTYY42 pKa = 11.12LNDD45 pKa = 4.07LDD47 pKa = 3.93THH49 pKa = 6.37EE50 pKa = 4.97PLVTFINTYY59 pKa = 10.24GFIYY63 pKa = 10.18EE64 pKa = 3.99QDD66 pKa = 3.28RR67 pKa = 11.84GNTIVGEE74 pKa = 4.08DD75 pKa = 3.34QLGKK79 pKa = 10.38KK80 pKa = 9.83RR81 pKa = 11.84EE82 pKa = 4.16AVTAAMVTLGCGPNLPSLGNVLGQLRR108 pKa = 11.84EE109 pKa = 4.0FQVTVRR115 pKa = 11.84KK116 pKa = 7.95TSSKK120 pKa = 9.45AEE122 pKa = 3.74EE123 pKa = 4.02MVFEE127 pKa = 4.11IVKK130 pKa = 9.73YY131 pKa = 8.79PRR133 pKa = 11.84IFRR136 pKa = 11.84GHH138 pKa = 5.43TLIQKK143 pKa = 10.21GLVCVSAEE151 pKa = 4.2KK152 pKa = 10.22FVKK155 pKa = 10.57SPGKK159 pKa = 9.4IQSGMDD165 pKa = 3.34YY166 pKa = 11.12LFIPTFLSVTYY177 pKa = 9.89CPAAIKK183 pKa = 9.84FQVPGPMLKK192 pKa = 8.94MRR194 pKa = 11.84SRR196 pKa = 11.84YY197 pKa = 6.47TQSLQLEE204 pKa = 4.25LMIRR208 pKa = 11.84ILCKK212 pKa = 9.79PDD214 pKa = 3.31SPLMKK219 pKa = 10.14VHH221 pKa = 6.4TPDD224 pKa = 3.67KK225 pKa = 10.25EE226 pKa = 4.28GRR228 pKa = 11.84GCLVSVWLHH237 pKa = 4.57VCNIFKK243 pKa = 10.67SGNKK247 pKa = 9.49NGSEE251 pKa = 3.74WQEE254 pKa = 3.32YY255 pKa = 7.21WMRR258 pKa = 11.84KK259 pKa = 7.19CANMQLEE266 pKa = 4.3VSIADD271 pKa = 3.19MWGPTIIIHH280 pKa = 6.41ARR282 pKa = 11.84GHH284 pKa = 5.71IPKK287 pKa = 9.82SAKK290 pKa = 10.39LFFGKK295 pKa = 10.44GGWSCHH301 pKa = 5.65PLHH304 pKa = 6.51EE305 pKa = 4.59VVPSVTKK312 pKa = 8.79TLWSVGCEE320 pKa = 3.51ITKK323 pKa = 10.6AKK325 pKa = 10.62AIIQEE330 pKa = 4.25SSISLLVEE338 pKa = 3.81TTDD341 pKa = 3.97IISPKK346 pKa = 10.19VKK348 pKa = 9.79ISSKK352 pKa = 9.77HH353 pKa = 4.19RR354 pKa = 11.84RR355 pKa = 11.84FVKK358 pKa = 10.78SNWGLFKK365 pKa = 10.08KK366 pKa = 9.01TKK368 pKa = 10.1SLPNLTEE375 pKa = 4.03LEE377 pKa = 4.23
Molecular weight: 42.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4893 |
44 |
2255 |
611.6 |
68.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.56 ± 0.995 | 1.962 ± 0.273 |
4.66 ± 0.485 | 4.925 ± 0.655 |
3.638 ± 0.357 | 5.702 ± 0.581 |
1.737 ± 0.392 | 7.766 ± 0.706 |
4.558 ± 0.908 | 11.179 ± 0.767 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.265 | 5.436 ± 0.477 |
5.109 ± 0.459 | 4.312 ± 0.493 |
4.68 ± 0.477 | 7.93 ± 0.533 |
7.46 ± 0.68 | 5.518 ± 0.731 |
1.165 ± 0.268 | 3.25 ± 0.497 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
