
Mycobacterium sp. 1245111.1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium; unclassified Mycobacterium
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
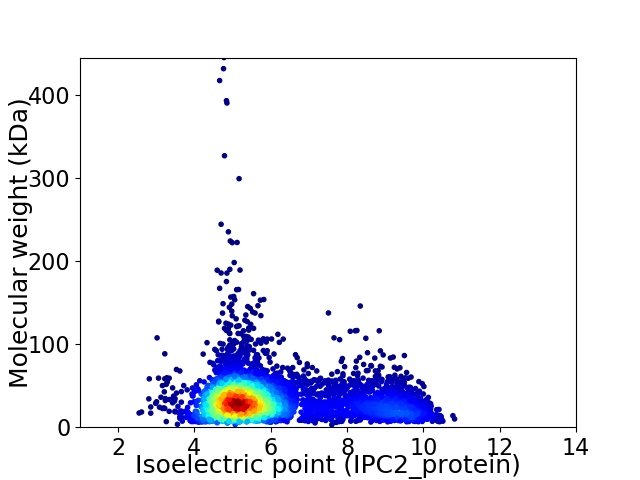
Virtual 2D-PAGE plot for 5178 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A3PIS4|A0A1A3PIS4_9MYCO RNA polymerase subunit sigma OS=Mycobacterium sp. 1245111.1 OX=1834073 GN=A5658_15790 PE=3 SV=1
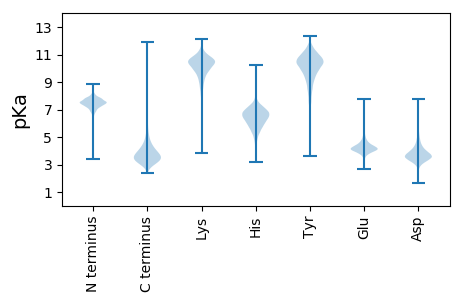
MM1 pKa = 7.84RR2 pKa = 11.84NVTWSSALPRR12 pKa = 11.84RR13 pKa = 11.84MRR15 pKa = 11.84SIGLAATAGAMAMAITAPHH34 pKa = 6.67ANADD38 pKa = 3.88PAYY41 pKa = 10.43QPALDD46 pKa = 3.81QLLEE50 pKa = 4.16AMQQAAAYY58 pKa = 7.73DD59 pKa = 3.62ASLPFATVDD68 pKa = 3.23SDD70 pKa = 3.7AMLMTGLQNTNYY82 pKa = 9.31EE83 pKa = 4.3LLSAGLSRR91 pKa = 11.84VTEE94 pKa = 4.12LFNGVFFQSPEE105 pKa = 3.85AVATDD110 pKa = 3.79AADD113 pKa = 3.21SRR115 pKa = 11.84QYY117 pKa = 11.11FQFFTPDD124 pKa = 2.61IYY126 pKa = 11.44YY127 pKa = 10.32HH128 pKa = 6.0ATAGLAPGATYY139 pKa = 10.82EE140 pKa = 4.14LTGTVGKK147 pKa = 8.72GTEE150 pKa = 3.89ALAIATEE157 pKa = 5.09AITGSTAVSKK167 pKa = 10.95EE168 pKa = 3.98SLEE171 pKa = 4.2LDD173 pKa = 2.91HH174 pKa = 7.05GLVVNPDD181 pKa = 2.9GSFTIDD187 pKa = 3.22IGPTEE192 pKa = 4.05PTGAVNFINDD202 pKa = 3.55TDD204 pKa = 3.55ATTNGDD210 pKa = 3.34ASLLIRR216 pKa = 11.84DD217 pKa = 4.34VLGDD221 pKa = 3.62WAQGPGSISIHH232 pKa = 6.11CVSDD236 pKa = 3.83CPAFFSIPSTGLFPGADD253 pKa = 3.14SSAAEE258 pKa = 4.18IGTLTGTSTPPITSVDD274 pKa = 3.69SLLTTLFTAFSNIVGPFNQEE294 pKa = 3.43NMGLAQVAGIDD305 pKa = 3.97LPPNTMSMLAPEE317 pKa = 4.36TSVFATGLPSADD329 pKa = 3.02VSGGNFDD336 pKa = 5.47LSPDD340 pKa = 3.21QALIVQVPDD349 pKa = 3.44VPSGYY354 pKa = 10.58SGIEE358 pKa = 3.74LMNVFGAALPFTLAQTTLNNTTAFADD384 pKa = 3.73PDD386 pKa = 4.06GYY388 pKa = 9.73TYY390 pKa = 11.32YY391 pKa = 10.81VVSATNPGVANWLDD405 pKa = 3.58SSGVSSGEE413 pKa = 3.3IFARR417 pKa = 11.84FEE419 pKa = 4.01NLPDD423 pKa = 3.98GVDD426 pKa = 3.51PTGLPVTTEE435 pKa = 4.14VVPVADD441 pKa = 3.59VANYY445 pKa = 10.25LPADD449 pKa = 3.98TPTVSPAEE457 pKa = 4.14YY458 pKa = 10.04AADD461 pKa = 3.34MSQRR465 pKa = 11.84VLSYY469 pKa = 10.88DD470 pKa = 3.41YY471 pKa = 11.63ALDD474 pKa = 3.87LSRR477 pKa = 11.84EE478 pKa = 4.08HH479 pKa = 6.93AQPDD483 pKa = 3.42WVLQEE488 pKa = 4.33LLLHH492 pKa = 6.5GFQGLMGTDD501 pKa = 2.72NFAAVFGSEE510 pKa = 3.83PSIPLDD516 pKa = 4.25LRR518 pKa = 11.84FTPALSPDD526 pKa = 3.65WATVDD531 pKa = 4.57HH532 pKa = 7.56DD533 pKa = 4.92LLTNPVASVEE543 pKa = 4.7AIFNTLPLAASDD555 pKa = 3.25ISLPTEE561 pKa = 3.74LAVGQTALSLLLPQQLGSLFNDD583 pKa = 5.4AILDD587 pKa = 3.97PNTGIIAGFLNARR600 pKa = 11.84DD601 pKa = 4.23DD602 pKa = 3.99LATAILTANNGFPTEE617 pKa = 4.16LGSVATAEE625 pKa = 4.18WANMPEE631 pKa = 4.5LVQSTASTLLTQLGALFGAA650 pKa = 4.94
MM1 pKa = 7.84RR2 pKa = 11.84NVTWSSALPRR12 pKa = 11.84RR13 pKa = 11.84MRR15 pKa = 11.84SIGLAATAGAMAMAITAPHH34 pKa = 6.67ANADD38 pKa = 3.88PAYY41 pKa = 10.43QPALDD46 pKa = 3.81QLLEE50 pKa = 4.16AMQQAAAYY58 pKa = 7.73DD59 pKa = 3.62ASLPFATVDD68 pKa = 3.23SDD70 pKa = 3.7AMLMTGLQNTNYY82 pKa = 9.31EE83 pKa = 4.3LLSAGLSRR91 pKa = 11.84VTEE94 pKa = 4.12LFNGVFFQSPEE105 pKa = 3.85AVATDD110 pKa = 3.79AADD113 pKa = 3.21SRR115 pKa = 11.84QYY117 pKa = 11.11FQFFTPDD124 pKa = 2.61IYY126 pKa = 11.44YY127 pKa = 10.32HH128 pKa = 6.0ATAGLAPGATYY139 pKa = 10.82EE140 pKa = 4.14LTGTVGKK147 pKa = 8.72GTEE150 pKa = 3.89ALAIATEE157 pKa = 5.09AITGSTAVSKK167 pKa = 10.95EE168 pKa = 3.98SLEE171 pKa = 4.2LDD173 pKa = 2.91HH174 pKa = 7.05GLVVNPDD181 pKa = 2.9GSFTIDD187 pKa = 3.22IGPTEE192 pKa = 4.05PTGAVNFINDD202 pKa = 3.55TDD204 pKa = 3.55ATTNGDD210 pKa = 3.34ASLLIRR216 pKa = 11.84DD217 pKa = 4.34VLGDD221 pKa = 3.62WAQGPGSISIHH232 pKa = 6.11CVSDD236 pKa = 3.83CPAFFSIPSTGLFPGADD253 pKa = 3.14SSAAEE258 pKa = 4.18IGTLTGTSTPPITSVDD274 pKa = 3.69SLLTTLFTAFSNIVGPFNQEE294 pKa = 3.43NMGLAQVAGIDD305 pKa = 3.97LPPNTMSMLAPEE317 pKa = 4.36TSVFATGLPSADD329 pKa = 3.02VSGGNFDD336 pKa = 5.47LSPDD340 pKa = 3.21QALIVQVPDD349 pKa = 3.44VPSGYY354 pKa = 10.58SGIEE358 pKa = 3.74LMNVFGAALPFTLAQTTLNNTTAFADD384 pKa = 3.73PDD386 pKa = 4.06GYY388 pKa = 9.73TYY390 pKa = 11.32YY391 pKa = 10.81VVSATNPGVANWLDD405 pKa = 3.58SSGVSSGEE413 pKa = 3.3IFARR417 pKa = 11.84FEE419 pKa = 4.01NLPDD423 pKa = 3.98GVDD426 pKa = 3.51PTGLPVTTEE435 pKa = 4.14VVPVADD441 pKa = 3.59VANYY445 pKa = 10.25LPADD449 pKa = 3.98TPTVSPAEE457 pKa = 4.14YY458 pKa = 10.04AADD461 pKa = 3.34MSQRR465 pKa = 11.84VLSYY469 pKa = 10.88DD470 pKa = 3.41YY471 pKa = 11.63ALDD474 pKa = 3.87LSRR477 pKa = 11.84EE478 pKa = 4.08HH479 pKa = 6.93AQPDD483 pKa = 3.42WVLQEE488 pKa = 4.33LLLHH492 pKa = 6.5GFQGLMGTDD501 pKa = 2.72NFAAVFGSEE510 pKa = 3.83PSIPLDD516 pKa = 4.25LRR518 pKa = 11.84FTPALSPDD526 pKa = 3.65WATVDD531 pKa = 4.57HH532 pKa = 7.56DD533 pKa = 4.92LLTNPVASVEE543 pKa = 4.7AIFNTLPLAASDD555 pKa = 3.25ISLPTEE561 pKa = 3.74LAVGQTALSLLLPQQLGSLFNDD583 pKa = 5.4AILDD587 pKa = 3.97PNTGIIAGFLNARR600 pKa = 11.84DD601 pKa = 4.23DD602 pKa = 3.99LATAILTANNGFPTEE617 pKa = 4.16LGSVATAEE625 pKa = 4.18WANMPEE631 pKa = 4.5LVQSTASTLLTQLGALFGAA650 pKa = 4.94
Molecular weight: 67.78 kDa
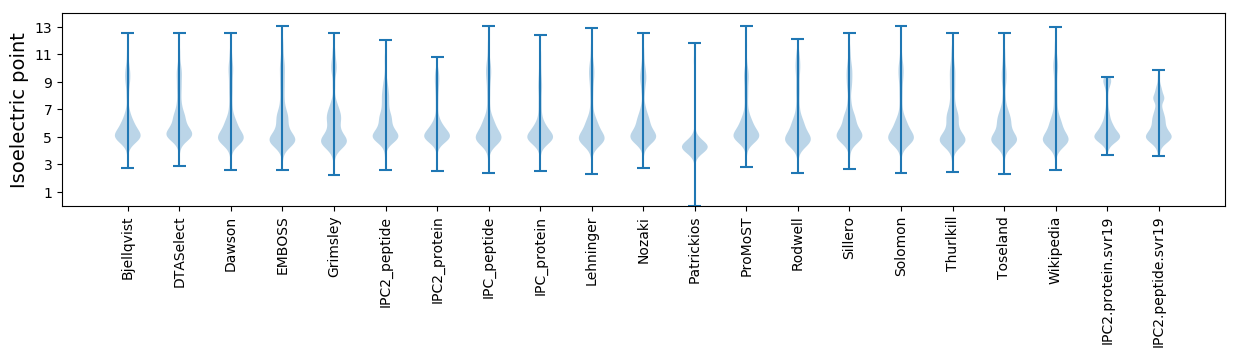
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A3Q2H2|A0A1A3Q2H2_9MYCO RNA polymerase subunit sigma OS=Mycobacterium sp. 1245111.1 OX=1834073 GN=A5658_02715 PE=3 SV=1
MM1 pKa = 7.32RR2 pKa = 11.84AGRR5 pKa = 11.84VFEE8 pKa = 4.13VTQEE12 pKa = 4.0VLHH15 pKa = 6.0ALIGRR20 pKa = 11.84VTDD23 pKa = 3.03IRR25 pKa = 11.84IRR27 pKa = 11.84LGLWLGLRR35 pKa = 11.84RR36 pKa = 11.84LLLWLGRR43 pKa = 11.84GLLLLRR49 pKa = 11.84WLRR52 pKa = 11.84WSVRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84LRR60 pKa = 11.84SLLGLWRR67 pKa = 11.84RR68 pKa = 11.84AQGLGGVTGAEE79 pKa = 4.08QRR81 pKa = 11.84WLRR84 pKa = 11.84RR85 pKa = 11.84SWRR88 pKa = 11.84GRR90 pKa = 11.84RR91 pKa = 11.84QWVLGRR97 pKa = 11.84HH98 pKa = 4.03IRR100 pKa = 11.84EE101 pKa = 3.95RR102 pKa = 11.84LRR104 pKa = 11.84GNAGGTGQWFRR115 pKa = 11.84HH116 pKa = 4.93
MM1 pKa = 7.32RR2 pKa = 11.84AGRR5 pKa = 11.84VFEE8 pKa = 4.13VTQEE12 pKa = 4.0VLHH15 pKa = 6.0ALIGRR20 pKa = 11.84VTDD23 pKa = 3.03IRR25 pKa = 11.84IRR27 pKa = 11.84LGLWLGLRR35 pKa = 11.84RR36 pKa = 11.84LLLWLGRR43 pKa = 11.84GLLLLRR49 pKa = 11.84WLRR52 pKa = 11.84WSVRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84LRR60 pKa = 11.84SLLGLWRR67 pKa = 11.84RR68 pKa = 11.84AQGLGGVTGAEE79 pKa = 4.08QRR81 pKa = 11.84WLRR84 pKa = 11.84RR85 pKa = 11.84SWRR88 pKa = 11.84GRR90 pKa = 11.84RR91 pKa = 11.84QWVLGRR97 pKa = 11.84HH98 pKa = 4.03IRR100 pKa = 11.84EE101 pKa = 3.95RR102 pKa = 11.84LRR104 pKa = 11.84GNAGGTGQWFRR115 pKa = 11.84HH116 pKa = 4.93
Molecular weight: 13.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1648677 |
27 |
4134 |
318.4 |
34.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.835 ± 0.049 | 0.813 ± 0.01 |
6.426 ± 0.027 | 5.215 ± 0.033 |
3.116 ± 0.02 | 8.736 ± 0.051 |
2.334 ± 0.016 | 4.388 ± 0.02 |
2.321 ± 0.022 | 9.741 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.066 ± 0.014 | 2.4 ± 0.018 |
5.792 ± 0.027 | 3.113 ± 0.017 |
7.038 ± 0.04 | 5.584 ± 0.024 |
5.924 ± 0.023 | 8.437 ± 0.031 |
1.532 ± 0.014 | 2.188 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
