
J-virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Jeilongvirus; Jun jeilongvirus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
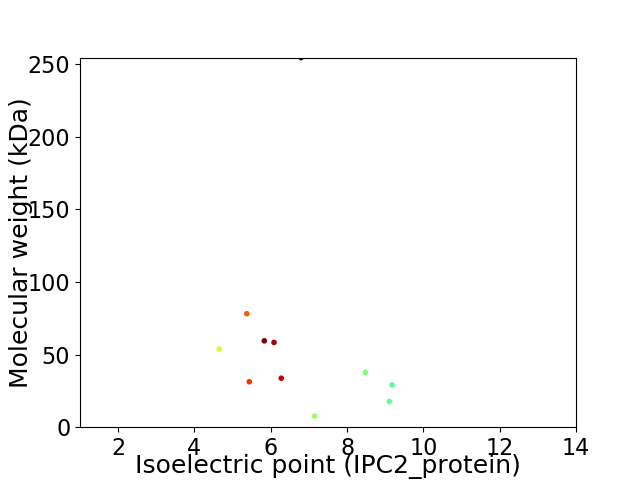
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q49HP2|Q49HP2_9MONO Nucleocapsid OS=J-virus OX=322067 GN=N PE=3 SV=1
MM1 pKa = 7.45NSYY4 pKa = 10.47SVQALEE10 pKa = 3.91QLVNDD15 pKa = 5.93GIKK18 pKa = 9.12TAQFFQKK25 pKa = 10.35NQEE28 pKa = 4.49NIQKK32 pKa = 9.13TYY34 pKa = 9.93GRR36 pKa = 11.84SAIGLPTTKK45 pKa = 10.2EE46 pKa = 3.95RR47 pKa = 11.84ISAWEE52 pKa = 3.84AVAEE56 pKa = 4.36TPYY59 pKa = 11.17GEE61 pKa = 4.13QIQLGGGNGEE71 pKa = 4.21DD72 pKa = 3.58QRR74 pKa = 11.84GEE76 pKa = 3.95QAEE79 pKa = 4.44GQDD82 pKa = 3.52NPGGHH87 pKa = 5.27GHH89 pKa = 6.65GGEE92 pKa = 4.64EE93 pKa = 4.48IPTGANPSALQLQGSYY109 pKa = 9.95NQVWDD114 pKa = 4.28GPNVATYY121 pKa = 10.71SGGEE125 pKa = 4.11GGDD128 pKa = 3.33TGNQHH133 pKa = 6.03GRR135 pKa = 11.84TDD137 pKa = 3.44SGSAADD143 pKa = 3.96TGGSNQSDD151 pKa = 3.82GSSTYY156 pKa = 9.67STVDD160 pKa = 3.02AGDD163 pKa = 3.85LRR165 pKa = 11.84QMMIFDD171 pKa = 4.86HH172 pKa = 6.04EE173 pKa = 4.35TSAIEE178 pKa = 4.36VGASKK183 pKa = 11.06NNTMKK188 pKa = 10.16IRR190 pKa = 11.84NATEE194 pKa = 3.58EE195 pKa = 4.02DD196 pKa = 3.5LGNVMSEE203 pKa = 4.29GTSKK207 pKa = 9.56IHH209 pKa = 6.15KK210 pKa = 9.2RR211 pKa = 11.84LRR213 pKa = 11.84GVAAMTDD220 pKa = 3.92PLPVPRR226 pKa = 11.84TTGGPVKK233 pKa = 10.46KK234 pKa = 10.31GIEE237 pKa = 4.11EE238 pKa = 4.42SSVSTILGGRR248 pKa = 11.84PTSGSGAIPNVHH260 pKa = 7.01PSLLLLPSQDD270 pKa = 2.88VHH272 pKa = 8.56AEE274 pKa = 4.05SAPKK278 pKa = 10.19CVQDD282 pKa = 3.5VSKK285 pKa = 9.74TGSTCQSSKK294 pKa = 11.38AEE296 pKa = 3.96QDD298 pKa = 3.3CSGIEE303 pKa = 4.03GKK305 pKa = 9.8IDD307 pKa = 3.82LLLGNLEE314 pKa = 4.23KK315 pKa = 10.28VTRR318 pKa = 11.84ALDD321 pKa = 3.38VLPEE325 pKa = 3.59IRR327 pKa = 11.84EE328 pKa = 4.05EE329 pKa = 3.63IRR331 pKa = 11.84NINKK335 pKa = 10.34KK336 pKa = 8.04ITNLSLGLSTVEE348 pKa = 4.54GYY350 pKa = 9.82IKK352 pKa = 10.95SMMIIIPGSGKK363 pKa = 9.55PNPGDD368 pKa = 3.57TPEE371 pKa = 4.17VNPDD375 pKa = 3.28LKK377 pKa = 11.17AVIGRR382 pKa = 11.84DD383 pKa = 3.25NTRR386 pKa = 11.84GLSEE390 pKa = 4.21VRR392 pKa = 11.84SQKK395 pKa = 10.38INLEE399 pKa = 4.14SLTDD403 pKa = 4.09DD404 pKa = 4.36PPSLGTIDD412 pKa = 3.72EE413 pKa = 4.35SHH415 pKa = 6.93IVRR418 pKa = 11.84DD419 pKa = 4.02LDD421 pKa = 3.83FTKK424 pKa = 11.05SNAANFAPTNRR435 pKa = 11.84FSSVMTMFNMIKK447 pKa = 10.45EE448 pKa = 4.2EE449 pKa = 4.04VKK451 pKa = 10.5DD452 pKa = 3.75YY453 pKa = 11.6NMRR456 pKa = 11.84AGLTRR461 pKa = 11.84WLEE464 pKa = 3.97DD465 pKa = 4.0AIEE468 pKa = 4.54RR469 pKa = 11.84LDD471 pKa = 3.92ADD473 pKa = 3.72DD474 pKa = 3.99VYY476 pKa = 11.11RR477 pKa = 11.84ALRR480 pKa = 11.84RR481 pKa = 11.84TLDD484 pKa = 3.25RR485 pKa = 11.84LAEE488 pKa = 4.5DD489 pKa = 5.09DD490 pKa = 4.53EE491 pKa = 6.64DD492 pKa = 6.11DD493 pKa = 4.86EE494 pKa = 6.0EE495 pKa = 4.62EE496 pKa = 5.09DD497 pKa = 3.87EE498 pKa = 4.33NN499 pKa = 4.99
MM1 pKa = 7.45NSYY4 pKa = 10.47SVQALEE10 pKa = 3.91QLVNDD15 pKa = 5.93GIKK18 pKa = 9.12TAQFFQKK25 pKa = 10.35NQEE28 pKa = 4.49NIQKK32 pKa = 9.13TYY34 pKa = 9.93GRR36 pKa = 11.84SAIGLPTTKK45 pKa = 10.2EE46 pKa = 3.95RR47 pKa = 11.84ISAWEE52 pKa = 3.84AVAEE56 pKa = 4.36TPYY59 pKa = 11.17GEE61 pKa = 4.13QIQLGGGNGEE71 pKa = 4.21DD72 pKa = 3.58QRR74 pKa = 11.84GEE76 pKa = 3.95QAEE79 pKa = 4.44GQDD82 pKa = 3.52NPGGHH87 pKa = 5.27GHH89 pKa = 6.65GGEE92 pKa = 4.64EE93 pKa = 4.48IPTGANPSALQLQGSYY109 pKa = 9.95NQVWDD114 pKa = 4.28GPNVATYY121 pKa = 10.71SGGEE125 pKa = 4.11GGDD128 pKa = 3.33TGNQHH133 pKa = 6.03GRR135 pKa = 11.84TDD137 pKa = 3.44SGSAADD143 pKa = 3.96TGGSNQSDD151 pKa = 3.82GSSTYY156 pKa = 9.67STVDD160 pKa = 3.02AGDD163 pKa = 3.85LRR165 pKa = 11.84QMMIFDD171 pKa = 4.86HH172 pKa = 6.04EE173 pKa = 4.35TSAIEE178 pKa = 4.36VGASKK183 pKa = 11.06NNTMKK188 pKa = 10.16IRR190 pKa = 11.84NATEE194 pKa = 3.58EE195 pKa = 4.02DD196 pKa = 3.5LGNVMSEE203 pKa = 4.29GTSKK207 pKa = 9.56IHH209 pKa = 6.15KK210 pKa = 9.2RR211 pKa = 11.84LRR213 pKa = 11.84GVAAMTDD220 pKa = 3.92PLPVPRR226 pKa = 11.84TTGGPVKK233 pKa = 10.46KK234 pKa = 10.31GIEE237 pKa = 4.11EE238 pKa = 4.42SSVSTILGGRR248 pKa = 11.84PTSGSGAIPNVHH260 pKa = 7.01PSLLLLPSQDD270 pKa = 2.88VHH272 pKa = 8.56AEE274 pKa = 4.05SAPKK278 pKa = 10.19CVQDD282 pKa = 3.5VSKK285 pKa = 9.74TGSTCQSSKK294 pKa = 11.38AEE296 pKa = 3.96QDD298 pKa = 3.3CSGIEE303 pKa = 4.03GKK305 pKa = 9.8IDD307 pKa = 3.82LLLGNLEE314 pKa = 4.23KK315 pKa = 10.28VTRR318 pKa = 11.84ALDD321 pKa = 3.38VLPEE325 pKa = 3.59IRR327 pKa = 11.84EE328 pKa = 4.05EE329 pKa = 3.63IRR331 pKa = 11.84NINKK335 pKa = 10.34KK336 pKa = 8.04ITNLSLGLSTVEE348 pKa = 4.54GYY350 pKa = 9.82IKK352 pKa = 10.95SMMIIIPGSGKK363 pKa = 9.55PNPGDD368 pKa = 3.57TPEE371 pKa = 4.17VNPDD375 pKa = 3.28LKK377 pKa = 11.17AVIGRR382 pKa = 11.84DD383 pKa = 3.25NTRR386 pKa = 11.84GLSEE390 pKa = 4.21VRR392 pKa = 11.84SQKK395 pKa = 10.38INLEE399 pKa = 4.14SLTDD403 pKa = 4.09DD404 pKa = 4.36PPSLGTIDD412 pKa = 3.72EE413 pKa = 4.35SHH415 pKa = 6.93IVRR418 pKa = 11.84DD419 pKa = 4.02LDD421 pKa = 3.83FTKK424 pKa = 11.05SNAANFAPTNRR435 pKa = 11.84FSSVMTMFNMIKK447 pKa = 10.45EE448 pKa = 4.2EE449 pKa = 4.04VKK451 pKa = 10.5DD452 pKa = 3.75YY453 pKa = 11.6NMRR456 pKa = 11.84AGLTRR461 pKa = 11.84WLEE464 pKa = 3.97DD465 pKa = 4.0AIEE468 pKa = 4.54RR469 pKa = 11.84LDD471 pKa = 3.92ADD473 pKa = 3.72DD474 pKa = 3.99VYY476 pKa = 11.11RR477 pKa = 11.84ALRR480 pKa = 11.84RR481 pKa = 11.84TLDD484 pKa = 3.25RR485 pKa = 11.84LAEE488 pKa = 4.5DD489 pKa = 5.09DD490 pKa = 4.53EE491 pKa = 6.64DD492 pKa = 6.11DD493 pKa = 4.86EE494 pKa = 6.0EE495 pKa = 4.62EE496 pKa = 5.09DD497 pKa = 3.87EE498 pKa = 4.33NN499 pKa = 4.99
Molecular weight: 53.68 kDa
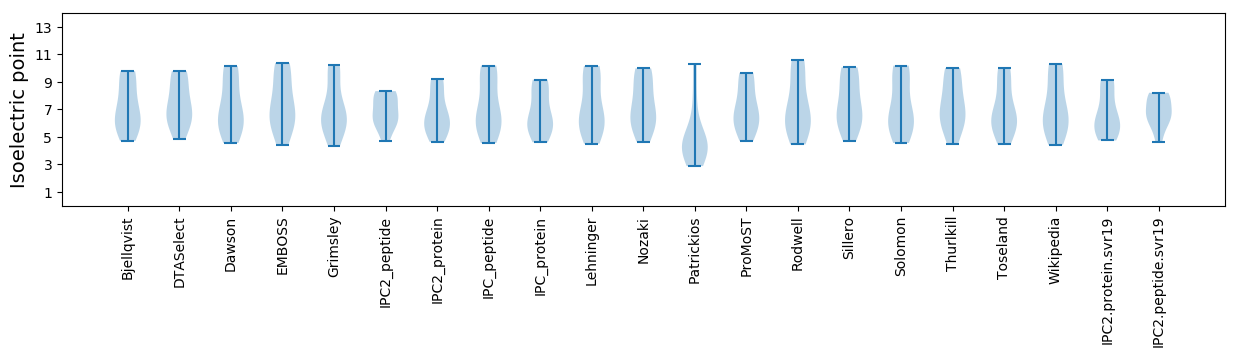
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q49HN5|Q49HN5_9MONO Small hydrophobic protein OS=J-virus OX=322067 GN=SH PE=4 SV=1
MM1 pKa = 7.49TSTVYY6 pKa = 10.64EE7 pKa = 4.46DD8 pKa = 4.08PEE10 pKa = 4.54SALSGYY16 pKa = 10.86EE17 pKa = 3.94SMRR20 pKa = 11.84SDD22 pKa = 3.96GARR25 pKa = 11.84GLPALPRR32 pKa = 11.84RR33 pKa = 11.84CKK35 pKa = 9.21TANTYY40 pKa = 10.42RR41 pKa = 11.84SARR44 pKa = 11.84RR45 pKa = 11.84VRR47 pKa = 11.84VVTRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VNNSVYY59 pKa = 9.72FVFIIVCLAILVAMAAYY76 pKa = 9.66VIIGIEE82 pKa = 3.96GLKK85 pKa = 7.47YY86 pKa = 9.22TRR88 pKa = 11.84YY89 pKa = 9.46NSKK92 pKa = 10.83DD93 pKa = 3.4VGPDD97 pKa = 3.05GSKK100 pKa = 10.43RR101 pKa = 11.84LEE103 pKa = 5.56DD104 pKa = 3.7IDD106 pKa = 4.34QKK108 pKa = 11.49LNQISSAVNTIMNALTYY125 pKa = 10.01SVPSVLSTYY134 pKa = 8.42RR135 pKa = 11.84TSLLNRR141 pKa = 11.84INHH144 pKa = 7.09LATEE148 pKa = 4.5LKK150 pKa = 10.26EE151 pKa = 3.9AARR154 pKa = 11.84MNNVDD159 pKa = 5.94LDD161 pKa = 4.44VKK163 pKa = 9.77WGSNRR168 pKa = 11.84TVLLKK173 pKa = 10.05TGSRR177 pKa = 11.84FHH179 pKa = 6.2QLNTRR184 pKa = 11.84EE185 pKa = 4.26LTTKK189 pKa = 8.88NTLVTSYY196 pKa = 10.7PRR198 pKa = 11.84MPTIIPKK205 pKa = 9.63VDD207 pKa = 3.16KK208 pKa = 10.23KK209 pKa = 10.83PPSFYY214 pKa = 10.47PLMKK218 pKa = 10.09VDD220 pKa = 4.9SDD222 pKa = 3.25QDD224 pKa = 3.68LNEE227 pKa = 4.11KK228 pKa = 9.79VKK230 pKa = 10.84AVTKK234 pKa = 9.97IFHH237 pKa = 6.85DD238 pKa = 3.54MSVTKK243 pKa = 10.44DD244 pKa = 3.67SQDD247 pKa = 3.23EE248 pKa = 4.72AIWNLNPASKK258 pKa = 10.76
MM1 pKa = 7.49TSTVYY6 pKa = 10.64EE7 pKa = 4.46DD8 pKa = 4.08PEE10 pKa = 4.54SALSGYY16 pKa = 10.86EE17 pKa = 3.94SMRR20 pKa = 11.84SDD22 pKa = 3.96GARR25 pKa = 11.84GLPALPRR32 pKa = 11.84RR33 pKa = 11.84CKK35 pKa = 9.21TANTYY40 pKa = 10.42RR41 pKa = 11.84SARR44 pKa = 11.84RR45 pKa = 11.84VRR47 pKa = 11.84VVTRR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84VNNSVYY59 pKa = 9.72FVFIIVCLAILVAMAAYY76 pKa = 9.66VIIGIEE82 pKa = 3.96GLKK85 pKa = 7.47YY86 pKa = 9.22TRR88 pKa = 11.84YY89 pKa = 9.46NSKK92 pKa = 10.83DD93 pKa = 3.4VGPDD97 pKa = 3.05GSKK100 pKa = 10.43RR101 pKa = 11.84LEE103 pKa = 5.56DD104 pKa = 3.7IDD106 pKa = 4.34QKK108 pKa = 11.49LNQISSAVNTIMNALTYY125 pKa = 10.01SVPSVLSTYY134 pKa = 8.42RR135 pKa = 11.84TSLLNRR141 pKa = 11.84INHH144 pKa = 7.09LATEE148 pKa = 4.5LKK150 pKa = 10.26EE151 pKa = 3.9AARR154 pKa = 11.84MNNVDD159 pKa = 5.94LDD161 pKa = 4.44VKK163 pKa = 9.77WGSNRR168 pKa = 11.84TVLLKK173 pKa = 10.05TGSRR177 pKa = 11.84FHH179 pKa = 6.2QLNTRR184 pKa = 11.84EE185 pKa = 4.26LTTKK189 pKa = 8.88NTLVTSYY196 pKa = 10.7PRR198 pKa = 11.84MPTIIPKK205 pKa = 9.63VDD207 pKa = 3.16KK208 pKa = 10.23KK209 pKa = 10.83PPSFYY214 pKa = 10.47PLMKK218 pKa = 10.09VDD220 pKa = 4.9SDD222 pKa = 3.25QDD224 pKa = 3.68LNEE227 pKa = 4.11KK228 pKa = 9.79VKK230 pKa = 10.84AVTKK234 pKa = 9.97IFHH237 pKa = 6.85DD238 pKa = 3.54MSVTKK243 pKa = 10.44DD244 pKa = 3.67SQDD247 pKa = 3.23EE248 pKa = 4.72AIWNLNPASKK258 pKa = 10.76
Molecular weight: 29.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5899 |
69 |
2204 |
536.3 |
60.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.628 ± 0.643 | 1.848 ± 0.303 |
5.509 ± 0.305 | 5.815 ± 0.458 |
3.357 ± 0.512 | 6.628 ± 1.178 |
2.0 ± 0.329 | 6.967 ± 0.515 |
5.882 ± 0.505 | 9.171 ± 0.807 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.068 ± 0.224 | 5.696 ± 0.29 |
4.357 ± 0.345 | 3.56 ± 0.498 |
5.035 ± 0.452 | 8.069 ± 0.417 |
6.306 ± 0.436 | 6.12 ± 0.387 |
1.17 ± 0.13 | 3.814 ± 0.479 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
