
Culex mononega-like virus 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
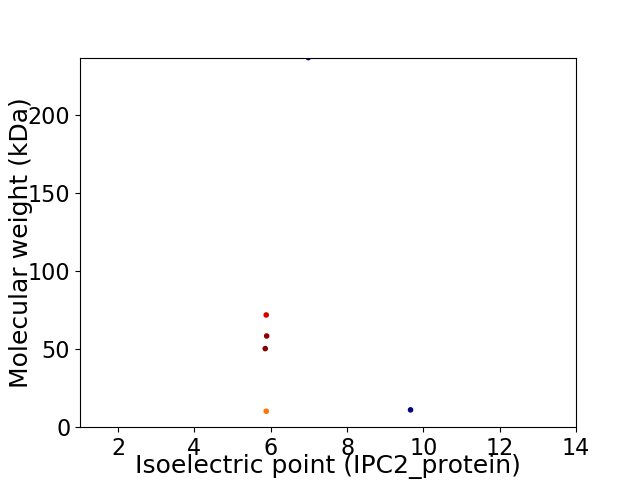
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RT68|A0A1Z2RT68_9VIRU Glycoprotein OS=Culex mononega-like virus 2 OX=2010272 PE=4 SV=1
MM1 pKa = 7.91KK2 pKa = 10.08MEE4 pKa = 3.75VHH6 pKa = 6.72LLVFLSFVALSAGFIAYY23 pKa = 8.55DD24 pKa = 3.82CNGSRR29 pKa = 11.84IQKK32 pKa = 5.62TTVSLVEE39 pKa = 4.29TPSCTFEE46 pKa = 4.41LEE48 pKa = 4.17NVTEE52 pKa = 4.33SEE54 pKa = 4.23VAVAVTQSALTQEE67 pKa = 4.13IPFYY71 pKa = 10.59RR72 pKa = 11.84CLIVAYY78 pKa = 9.28HH79 pKa = 6.35HH80 pKa = 6.77IWRR83 pKa = 11.84CGSVIDD89 pKa = 4.39TNVQGGDD96 pKa = 3.4YY97 pKa = 11.03AEE99 pKa = 4.79VIRR102 pKa = 11.84TTKK105 pKa = 10.4SEE107 pKa = 4.11CEE109 pKa = 3.49EE110 pKa = 4.24MINHH114 pKa = 6.93KK115 pKa = 10.08KK116 pKa = 10.6YY117 pKa = 9.45KK118 pKa = 9.51TIGGNSVSIDD128 pKa = 3.91LPPGGRR134 pKa = 11.84TSFSYY139 pKa = 10.41TSWGQVKK146 pKa = 10.67DD147 pKa = 3.6SGSCVAGPILYY158 pKa = 10.48SGGTSYY164 pKa = 11.12DD165 pKa = 2.95QHH167 pKa = 5.68TRR169 pKa = 11.84NTRR172 pKa = 11.84LEE174 pKa = 3.75ILYY177 pKa = 8.78TRR179 pKa = 11.84GVGRR183 pKa = 11.84VAVEE187 pKa = 4.12KK188 pKa = 10.69KK189 pKa = 10.64SLIMPNGMQCNVALEE204 pKa = 4.17EE205 pKa = 5.04CEE207 pKa = 3.9LADD210 pKa = 3.71YY211 pKa = 10.49GQVFWKK217 pKa = 10.45QPVPSCQNEE226 pKa = 4.09VEE228 pKa = 4.36EE229 pKa = 4.44QSLVYY234 pKa = 10.16KK235 pKa = 10.91GPATLITNQNTSEE248 pKa = 4.23RR249 pKa = 11.84FIQVSFSGHH258 pKa = 5.81HH259 pKa = 5.22FQIKK263 pKa = 10.46LEE265 pKa = 4.18DD266 pKa = 3.52RR267 pKa = 11.84TTYY270 pKa = 10.06ICGFRR275 pKa = 11.84SFFTEE280 pKa = 3.98HH281 pKa = 6.57PKK283 pKa = 10.92LYY285 pKa = 9.02ITLLDD290 pKa = 3.87LSYY293 pKa = 11.27PDD295 pKa = 5.21FPLKK299 pKa = 10.78EE300 pKa = 4.08NTGTDD305 pKa = 3.88VNMMNYY311 pKa = 9.47INSKK315 pKa = 8.94LVYY318 pKa = 9.84SMRR321 pKa = 11.84HH322 pKa = 4.85IKK324 pKa = 8.98EE325 pKa = 3.88QVMTLYY331 pKa = 10.91RR332 pKa = 11.84LFEE335 pKa = 4.3LEE337 pKa = 3.86RR338 pKa = 11.84CLMQNRR344 pKa = 11.84ITSNLLTLAILSPRR358 pKa = 11.84EE359 pKa = 3.51FAYY362 pKa = 10.18QYY364 pKa = 10.3YY365 pKa = 10.35GEE367 pKa = 4.56PGYY370 pKa = 8.47TAVVRR375 pKa = 11.84GEE377 pKa = 4.15VVHH380 pKa = 7.03IARR383 pKa = 11.84CTPVPVTPRR392 pKa = 11.84KK393 pKa = 8.5TEE395 pKa = 3.64LCYY398 pKa = 11.04NEE400 pKa = 5.24LPVSFNNKK408 pKa = 9.16SLFMTPRR415 pKa = 11.84SRR417 pKa = 11.84ILMEE421 pKa = 4.4IGTVVEE427 pKa = 4.73CATDD431 pKa = 3.16VGPQFKK437 pKa = 8.67TANRR441 pKa = 11.84WITMVSHH448 pKa = 6.51GLISVEE454 pKa = 4.04KK455 pKa = 9.57PKK457 pKa = 10.46IITPDD462 pKa = 4.4PITYY466 pKa = 10.04HH467 pKa = 6.68FEE469 pKa = 3.99TLEE472 pKa = 4.11DD473 pKa = 3.73MLTGGLYY480 pKa = 9.95TKK482 pKa = 8.99EE483 pKa = 4.04TIANYY488 pKa = 8.18QQILTSPMEE497 pKa = 3.93EE498 pKa = 4.43SILSSRR504 pKa = 11.84VTTAIRR510 pKa = 11.84GGEE513 pKa = 4.3SLPSGYY519 pKa = 9.15PASNIFSSYY528 pKa = 10.44DD529 pKa = 3.22IEE531 pKa = 4.19RR532 pKa = 11.84LRR534 pKa = 11.84VKK536 pKa = 9.22VTSGLVHH543 pKa = 6.49TMDD546 pKa = 4.18KK547 pKa = 11.04LLVLGNWFSVFVMIGWAINTLISIINCGLNYY578 pKa = 10.5YY579 pKa = 7.93VVKK582 pKa = 10.42PEE584 pKa = 3.84TSGIFALLTCCFSAIFQVVKK604 pKa = 10.55NRR606 pKa = 11.84QLGNPEE612 pKa = 3.77EE613 pKa = 4.44TKK615 pKa = 10.12TLSAVVQIDD624 pKa = 3.51AEE626 pKa = 4.41GRR628 pKa = 11.84EE629 pKa = 4.3EE630 pKa = 3.87PSHH633 pKa = 6.03NKK635 pKa = 9.51FYY637 pKa = 11.02KK638 pKa = 10.4
MM1 pKa = 7.91KK2 pKa = 10.08MEE4 pKa = 3.75VHH6 pKa = 6.72LLVFLSFVALSAGFIAYY23 pKa = 8.55DD24 pKa = 3.82CNGSRR29 pKa = 11.84IQKK32 pKa = 5.62TTVSLVEE39 pKa = 4.29TPSCTFEE46 pKa = 4.41LEE48 pKa = 4.17NVTEE52 pKa = 4.33SEE54 pKa = 4.23VAVAVTQSALTQEE67 pKa = 4.13IPFYY71 pKa = 10.59RR72 pKa = 11.84CLIVAYY78 pKa = 9.28HH79 pKa = 6.35HH80 pKa = 6.77IWRR83 pKa = 11.84CGSVIDD89 pKa = 4.39TNVQGGDD96 pKa = 3.4YY97 pKa = 11.03AEE99 pKa = 4.79VIRR102 pKa = 11.84TTKK105 pKa = 10.4SEE107 pKa = 4.11CEE109 pKa = 3.49EE110 pKa = 4.24MINHH114 pKa = 6.93KK115 pKa = 10.08KK116 pKa = 10.6YY117 pKa = 9.45KK118 pKa = 9.51TIGGNSVSIDD128 pKa = 3.91LPPGGRR134 pKa = 11.84TSFSYY139 pKa = 10.41TSWGQVKK146 pKa = 10.67DD147 pKa = 3.6SGSCVAGPILYY158 pKa = 10.48SGGTSYY164 pKa = 11.12DD165 pKa = 2.95QHH167 pKa = 5.68TRR169 pKa = 11.84NTRR172 pKa = 11.84LEE174 pKa = 3.75ILYY177 pKa = 8.78TRR179 pKa = 11.84GVGRR183 pKa = 11.84VAVEE187 pKa = 4.12KK188 pKa = 10.69KK189 pKa = 10.64SLIMPNGMQCNVALEE204 pKa = 4.17EE205 pKa = 5.04CEE207 pKa = 3.9LADD210 pKa = 3.71YY211 pKa = 10.49GQVFWKK217 pKa = 10.45QPVPSCQNEE226 pKa = 4.09VEE228 pKa = 4.36EE229 pKa = 4.44QSLVYY234 pKa = 10.16KK235 pKa = 10.91GPATLITNQNTSEE248 pKa = 4.23RR249 pKa = 11.84FIQVSFSGHH258 pKa = 5.81HH259 pKa = 5.22FQIKK263 pKa = 10.46LEE265 pKa = 4.18DD266 pKa = 3.52RR267 pKa = 11.84TTYY270 pKa = 10.06ICGFRR275 pKa = 11.84SFFTEE280 pKa = 3.98HH281 pKa = 6.57PKK283 pKa = 10.92LYY285 pKa = 9.02ITLLDD290 pKa = 3.87LSYY293 pKa = 11.27PDD295 pKa = 5.21FPLKK299 pKa = 10.78EE300 pKa = 4.08NTGTDD305 pKa = 3.88VNMMNYY311 pKa = 9.47INSKK315 pKa = 8.94LVYY318 pKa = 9.84SMRR321 pKa = 11.84HH322 pKa = 4.85IKK324 pKa = 8.98EE325 pKa = 3.88QVMTLYY331 pKa = 10.91RR332 pKa = 11.84LFEE335 pKa = 4.3LEE337 pKa = 3.86RR338 pKa = 11.84CLMQNRR344 pKa = 11.84ITSNLLTLAILSPRR358 pKa = 11.84EE359 pKa = 3.51FAYY362 pKa = 10.18QYY364 pKa = 10.3YY365 pKa = 10.35GEE367 pKa = 4.56PGYY370 pKa = 8.47TAVVRR375 pKa = 11.84GEE377 pKa = 4.15VVHH380 pKa = 7.03IARR383 pKa = 11.84CTPVPVTPRR392 pKa = 11.84KK393 pKa = 8.5TEE395 pKa = 3.64LCYY398 pKa = 11.04NEE400 pKa = 5.24LPVSFNNKK408 pKa = 9.16SLFMTPRR415 pKa = 11.84SRR417 pKa = 11.84ILMEE421 pKa = 4.4IGTVVEE427 pKa = 4.73CATDD431 pKa = 3.16VGPQFKK437 pKa = 8.67TANRR441 pKa = 11.84WITMVSHH448 pKa = 6.51GLISVEE454 pKa = 4.04KK455 pKa = 9.57PKK457 pKa = 10.46IITPDD462 pKa = 4.4PITYY466 pKa = 10.04HH467 pKa = 6.68FEE469 pKa = 3.99TLEE472 pKa = 4.11DD473 pKa = 3.73MLTGGLYY480 pKa = 9.95TKK482 pKa = 8.99EE483 pKa = 4.04TIANYY488 pKa = 8.18QQILTSPMEE497 pKa = 3.93EE498 pKa = 4.43SILSSRR504 pKa = 11.84VTTAIRR510 pKa = 11.84GGEE513 pKa = 4.3SLPSGYY519 pKa = 9.15PASNIFSSYY528 pKa = 10.44DD529 pKa = 3.22IEE531 pKa = 4.19RR532 pKa = 11.84LRR534 pKa = 11.84VKK536 pKa = 9.22VTSGLVHH543 pKa = 6.49TMDD546 pKa = 4.18KK547 pKa = 11.04LLVLGNWFSVFVMIGWAINTLISIINCGLNYY578 pKa = 10.5YY579 pKa = 7.93VVKK582 pKa = 10.42PEE584 pKa = 3.84TSGIFALLTCCFSAIFQVVKK604 pKa = 10.55NRR606 pKa = 11.84QLGNPEE612 pKa = 3.77EE613 pKa = 4.44TKK615 pKa = 10.12TLSAVVQIDD624 pKa = 3.51AEE626 pKa = 4.41GRR628 pKa = 11.84EE629 pKa = 4.3EE630 pKa = 3.87PSHH633 pKa = 6.03NKK635 pKa = 9.51FYY637 pKa = 11.02KK638 pKa = 10.4
Molecular weight: 71.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RT27|A0A1Z2RT27_9VIRU Uncharacterized protein OS=Culex mononega-like virus 2 OX=2010272 PE=4 SV=1
MM1 pKa = 7.48NYY3 pKa = 7.67ITPTILVAVIVIAHH17 pKa = 5.68TVAIRR22 pKa = 11.84SLSYY26 pKa = 11.64DD27 pKa = 3.05MTDD30 pKa = 3.05RR31 pKa = 11.84VPVIALINDD40 pKa = 3.49HH41 pKa = 6.88ASWFMKK47 pKa = 9.54LTVSPVTINFYY58 pKa = 10.09ATLTSFFATLALMGVVAKK76 pKa = 10.36NWRR79 pKa = 11.84AYY81 pKa = 10.3VRR83 pKa = 11.84DD84 pKa = 3.37IQRR87 pKa = 11.84GVVRR91 pKa = 11.84AEE93 pKa = 3.54VLMRR97 pKa = 11.84SAA99 pKa = 4.92
MM1 pKa = 7.48NYY3 pKa = 7.67ITPTILVAVIVIAHH17 pKa = 5.68TVAIRR22 pKa = 11.84SLSYY26 pKa = 11.64DD27 pKa = 3.05MTDD30 pKa = 3.05RR31 pKa = 11.84VPVIALINDD40 pKa = 3.49HH41 pKa = 6.88ASWFMKK47 pKa = 9.54LTVSPVTINFYY58 pKa = 10.09ATLTSFFATLALMGVVAKK76 pKa = 10.36NWRR79 pKa = 11.84AYY81 pKa = 10.3VRR83 pKa = 11.84DD84 pKa = 3.37IQRR87 pKa = 11.84GVVRR91 pKa = 11.84AEE93 pKa = 3.54VLMRR97 pKa = 11.84SAA99 pKa = 4.92
Molecular weight: 11.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3855 |
86 |
2079 |
642.5 |
73.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.811 ± 0.572 | 2.387 ± 0.232 |
4.929 ± 0.819 | 6.978 ± 0.467 |
4.099 ± 0.248 | 5.785 ± 0.421 |
2.464 ± 0.144 | 6.304 ± 0.426 |
5.759 ± 0.662 | 9.183 ± 0.265 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.32 ± 0.178 | 4.047 ± 0.438 |
4.202 ± 0.244 | 3.398 ± 0.156 |
5.914 ± 0.436 | 6.848 ± 0.69 |
6.667 ± 0.887 | 6.693 ± 0.566 |
1.167 ± 0.162 | 4.047 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
