
Pseudomonas pohangensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
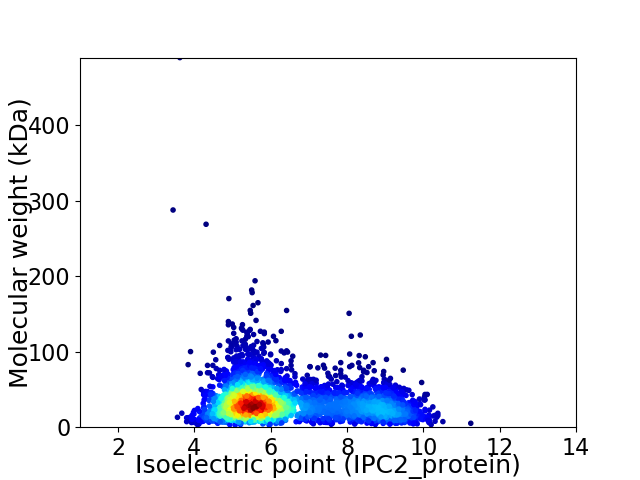
Virtual 2D-PAGE plot for 3431 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H2HUI6|A0A1H2HUI6_9PSED Glucose-6-phosphate 1-dehydrogenase OS=Pseudomonas pohangensis OX=364197 GN=zwf PE=3 SV=1
MM1 pKa = 7.53NKK3 pKa = 8.88TNNATPFAKK12 pKa = 9.81TPLVIAIACAGWMSAAQAGEE32 pKa = 4.05ILDD35 pKa = 4.19LSYY38 pKa = 8.3TTGDD42 pKa = 3.49GYY44 pKa = 10.86ILIDD48 pKa = 3.8ADD50 pKa = 4.14EE51 pKa = 4.45EE52 pKa = 4.78VVAPGVKK59 pKa = 10.07AVTTDD64 pKa = 3.41LNNNDD69 pKa = 3.24FVGNGFSPSGDD80 pKa = 4.1PNCLMASSPATCDD93 pKa = 3.21SPKK96 pKa = 10.83NSGKK100 pKa = 10.4RR101 pKa = 11.84IKK103 pKa = 10.97NNLTGRR109 pKa = 11.84GAFEE113 pKa = 3.58HH114 pKa = 6.5VYY116 pKa = 10.31NVKK119 pKa = 9.82TSGGTTEE126 pKa = 3.81YY127 pKa = 11.09LNYY130 pKa = 10.91GKK132 pKa = 8.43ITNRR136 pKa = 11.84TGARR140 pKa = 11.84LLGYY144 pKa = 8.39QVIVGTGTGANFTPASASDD163 pKa = 3.69QAGLLAMDD171 pKa = 4.2NVLPLSGQATSWPGTTQAEE190 pKa = 4.69GQSPLQRR197 pKa = 11.84TWLPEE202 pKa = 3.71GLFGTKK208 pKa = 8.37QDD210 pKa = 3.3MSAGYY215 pKa = 9.22FSPDD219 pKa = 2.69NAGFFFEE226 pKa = 4.75TVGTDD231 pKa = 3.22TLSADD236 pKa = 3.23GMFANTVYY244 pKa = 10.89SQFFGDD250 pKa = 3.91GLLTRR255 pKa = 11.84GQQPQALFLDD265 pKa = 4.0NLDD268 pKa = 4.55PDD270 pKa = 3.94VEE272 pKa = 4.35DD273 pKa = 6.49DD274 pKa = 3.33IVYY277 pKa = 9.72WKK279 pKa = 10.79AGNLWLDD286 pKa = 3.4AAGTVQDD293 pKa = 4.21TATVDD298 pKa = 3.4ALLATPGYY306 pKa = 10.12YY307 pKa = 10.13VDD309 pKa = 4.56TIEE312 pKa = 6.36DD313 pKa = 3.79LTNLNINFSMDD324 pKa = 3.14IGDD327 pKa = 3.88LTAGQFTVRR336 pKa = 11.84YY337 pKa = 8.79VPVFSPIVNAASSDD351 pKa = 3.82YY352 pKa = 10.74QLAVANSLDD361 pKa = 3.68ATQIPYY367 pKa = 10.85LFFDD371 pKa = 3.62QNTAAGATPTPTAAYY386 pKa = 10.71YY387 pKa = 9.65EE388 pKa = 4.45FQDD391 pKa = 4.33IMNAFDD397 pKa = 4.72ALDD400 pKa = 3.95PAAQSQALGSMGTSYY415 pKa = 10.95LRR417 pKa = 11.84NYY419 pKa = 7.31GTQGLLMGRR428 pKa = 11.84GTLEE432 pKa = 3.82AVQQHH437 pKa = 5.21LQEE440 pKa = 4.33NRR442 pKa = 11.84ISSLTPSSVSAEE454 pKa = 3.98TLANQLMGTPGAATLDD470 pKa = 3.69QVVGQPITSTADD482 pKa = 3.09LAMALDD488 pKa = 4.64DD489 pKa = 4.59NGNTASVALNEE500 pKa = 4.34TTSTFISGSASEE512 pKa = 4.83GNLDD516 pKa = 3.68NSNNGTGADD525 pKa = 3.61YY526 pKa = 10.87SAYY529 pKa = 10.35SLTVGLDD536 pKa = 2.92HH537 pKa = 7.11YY538 pKa = 11.38LRR540 pKa = 11.84DD541 pKa = 3.75QLRR544 pKa = 11.84VGAALGYY551 pKa = 9.32GHH553 pKa = 6.85NEE555 pKa = 3.44GDD557 pKa = 4.05VNDD560 pKa = 4.0NYY562 pKa = 11.68GNLDD566 pKa = 3.65LDD568 pKa = 4.77GYY570 pKa = 11.12SLTTFVSYY578 pKa = 11.01GGATGLFTDD587 pKa = 4.17VLLGYY592 pKa = 10.17SWLDD596 pKa = 3.18YY597 pKa = 11.65DD598 pKa = 4.6NDD600 pKa = 3.65RR601 pKa = 11.84KK602 pKa = 10.64INIGSEE608 pKa = 3.76QRR610 pKa = 11.84KK611 pKa = 9.47AKK613 pKa = 10.81SNTDD617 pKa = 2.88GDD619 pKa = 4.1LSSFAVRR626 pKa = 11.84SGYY629 pKa = 10.06NFEE632 pKa = 5.15LGPVIAGPSLRR643 pKa = 11.84YY644 pKa = 9.62QYY646 pKa = 11.49LDD648 pKa = 4.1LSVDD652 pKa = 4.04DD653 pKa = 4.35YY654 pKa = 10.89TEE656 pKa = 4.1KK657 pKa = 10.97DD658 pKa = 3.17AGVLNMHH665 pKa = 6.46VDD667 pKa = 3.49DD668 pKa = 4.66MSYY671 pKa = 11.35DD672 pKa = 3.45SSTLGLGGQMTMPIALEE689 pKa = 4.15SGFLRR694 pKa = 11.84PYY696 pKa = 11.27AEE698 pKa = 3.96AHH700 pKa = 5.08WVKK703 pKa = 10.37EE704 pKa = 4.03FQDD707 pKa = 4.63DD708 pKa = 3.72SSQVDD713 pKa = 3.37TSFVSGVVPFQTPIDD728 pKa = 4.52AYY730 pKa = 10.59DD731 pKa = 3.57SDD733 pKa = 4.52YY734 pKa = 8.73MTVGAGVEE742 pKa = 4.41TTFMASGMPASLSVNYY758 pKa = 10.35DD759 pKa = 3.32GVVSNSDD766 pKa = 3.29YY767 pKa = 11.16SDD769 pKa = 3.28NRR771 pKa = 11.84VSLDD775 pKa = 3.28LRR777 pKa = 11.84LAFF780 pKa = 5.09
MM1 pKa = 7.53NKK3 pKa = 8.88TNNATPFAKK12 pKa = 9.81TPLVIAIACAGWMSAAQAGEE32 pKa = 4.05ILDD35 pKa = 4.19LSYY38 pKa = 8.3TTGDD42 pKa = 3.49GYY44 pKa = 10.86ILIDD48 pKa = 3.8ADD50 pKa = 4.14EE51 pKa = 4.45EE52 pKa = 4.78VVAPGVKK59 pKa = 10.07AVTTDD64 pKa = 3.41LNNNDD69 pKa = 3.24FVGNGFSPSGDD80 pKa = 4.1PNCLMASSPATCDD93 pKa = 3.21SPKK96 pKa = 10.83NSGKK100 pKa = 10.4RR101 pKa = 11.84IKK103 pKa = 10.97NNLTGRR109 pKa = 11.84GAFEE113 pKa = 3.58HH114 pKa = 6.5VYY116 pKa = 10.31NVKK119 pKa = 9.82TSGGTTEE126 pKa = 3.81YY127 pKa = 11.09LNYY130 pKa = 10.91GKK132 pKa = 8.43ITNRR136 pKa = 11.84TGARR140 pKa = 11.84LLGYY144 pKa = 8.39QVIVGTGTGANFTPASASDD163 pKa = 3.69QAGLLAMDD171 pKa = 4.2NVLPLSGQATSWPGTTQAEE190 pKa = 4.69GQSPLQRR197 pKa = 11.84TWLPEE202 pKa = 3.71GLFGTKK208 pKa = 8.37QDD210 pKa = 3.3MSAGYY215 pKa = 9.22FSPDD219 pKa = 2.69NAGFFFEE226 pKa = 4.75TVGTDD231 pKa = 3.22TLSADD236 pKa = 3.23GMFANTVYY244 pKa = 10.89SQFFGDD250 pKa = 3.91GLLTRR255 pKa = 11.84GQQPQALFLDD265 pKa = 4.0NLDD268 pKa = 4.55PDD270 pKa = 3.94VEE272 pKa = 4.35DD273 pKa = 6.49DD274 pKa = 3.33IVYY277 pKa = 9.72WKK279 pKa = 10.79AGNLWLDD286 pKa = 3.4AAGTVQDD293 pKa = 4.21TATVDD298 pKa = 3.4ALLATPGYY306 pKa = 10.12YY307 pKa = 10.13VDD309 pKa = 4.56TIEE312 pKa = 6.36DD313 pKa = 3.79LTNLNINFSMDD324 pKa = 3.14IGDD327 pKa = 3.88LTAGQFTVRR336 pKa = 11.84YY337 pKa = 8.79VPVFSPIVNAASSDD351 pKa = 3.82YY352 pKa = 10.74QLAVANSLDD361 pKa = 3.68ATQIPYY367 pKa = 10.85LFFDD371 pKa = 3.62QNTAAGATPTPTAAYY386 pKa = 10.71YY387 pKa = 9.65EE388 pKa = 4.45FQDD391 pKa = 4.33IMNAFDD397 pKa = 4.72ALDD400 pKa = 3.95PAAQSQALGSMGTSYY415 pKa = 10.95LRR417 pKa = 11.84NYY419 pKa = 7.31GTQGLLMGRR428 pKa = 11.84GTLEE432 pKa = 3.82AVQQHH437 pKa = 5.21LQEE440 pKa = 4.33NRR442 pKa = 11.84ISSLTPSSVSAEE454 pKa = 3.98TLANQLMGTPGAATLDD470 pKa = 3.69QVVGQPITSTADD482 pKa = 3.09LAMALDD488 pKa = 4.64DD489 pKa = 4.59NGNTASVALNEE500 pKa = 4.34TTSTFISGSASEE512 pKa = 4.83GNLDD516 pKa = 3.68NSNNGTGADD525 pKa = 3.61YY526 pKa = 10.87SAYY529 pKa = 10.35SLTVGLDD536 pKa = 2.92HH537 pKa = 7.11YY538 pKa = 11.38LRR540 pKa = 11.84DD541 pKa = 3.75QLRR544 pKa = 11.84VGAALGYY551 pKa = 9.32GHH553 pKa = 6.85NEE555 pKa = 3.44GDD557 pKa = 4.05VNDD560 pKa = 4.0NYY562 pKa = 11.68GNLDD566 pKa = 3.65LDD568 pKa = 4.77GYY570 pKa = 11.12SLTTFVSYY578 pKa = 11.01GGATGLFTDD587 pKa = 4.17VLLGYY592 pKa = 10.17SWLDD596 pKa = 3.18YY597 pKa = 11.65DD598 pKa = 4.6NDD600 pKa = 3.65RR601 pKa = 11.84KK602 pKa = 10.64INIGSEE608 pKa = 3.76QRR610 pKa = 11.84KK611 pKa = 9.47AKK613 pKa = 10.81SNTDD617 pKa = 2.88GDD619 pKa = 4.1LSSFAVRR626 pKa = 11.84SGYY629 pKa = 10.06NFEE632 pKa = 5.15LGPVIAGPSLRR643 pKa = 11.84YY644 pKa = 9.62QYY646 pKa = 11.49LDD648 pKa = 4.1LSVDD652 pKa = 4.04DD653 pKa = 4.35YY654 pKa = 10.89TEE656 pKa = 4.1KK657 pKa = 10.97DD658 pKa = 3.17AGVLNMHH665 pKa = 6.46VDD667 pKa = 3.49DD668 pKa = 4.66MSYY671 pKa = 11.35DD672 pKa = 3.45SSTLGLGGQMTMPIALEE689 pKa = 4.15SGFLRR694 pKa = 11.84PYY696 pKa = 11.27AEE698 pKa = 3.96AHH700 pKa = 5.08WVKK703 pKa = 10.37EE704 pKa = 4.03FQDD707 pKa = 4.63DD708 pKa = 3.72SSQVDD713 pKa = 3.37TSFVSGVVPFQTPIDD728 pKa = 4.52AYY730 pKa = 10.59DD731 pKa = 3.57SDD733 pKa = 4.52YY734 pKa = 8.73MTVGAGVEE742 pKa = 4.41TTFMASGMPASLSVNYY758 pKa = 10.35DD759 pKa = 3.32GVVSNSDD766 pKa = 3.29YY767 pKa = 11.16SDD769 pKa = 3.28NRR771 pKa = 11.84VSLDD775 pKa = 3.28LRR777 pKa = 11.84LAFF780 pKa = 5.09
Molecular weight: 82.94 kDa
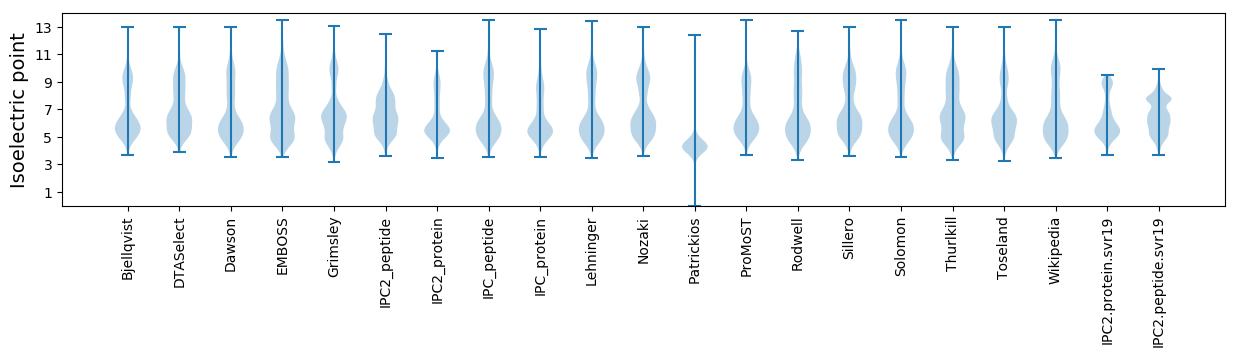
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2HY28|A0A1H2HY28_9PSED Ribosomal RNA small subunit methyltransferase G OS=Pseudomonas pohangensis OX=364197 GN=rsmG PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.24NGRR28 pKa = 11.84LVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.24NGRR28 pKa = 11.84LVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1117736 |
29 |
4753 |
325.8 |
35.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.014 ± 0.048 | 1.103 ± 0.014 |
5.227 ± 0.039 | 5.744 ± 0.045 |
3.683 ± 0.027 | 7.85 ± 0.044 |
2.197 ± 0.02 | 4.891 ± 0.03 |
3.554 ± 0.036 | 12.123 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.408 ± 0.019 | 3.055 ± 0.027 |
4.862 ± 0.029 | 4.853 ± 0.037 |
6.284 ± 0.047 | 5.821 ± 0.031 |
4.567 ± 0.056 | 6.751 ± 0.035 |
1.456 ± 0.018 | 2.559 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
