
Suicoccus acidiformans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Aerococcaceae; Suicoccus
Average proteome isoelectric point is 5.63
Get precalculated fractions of proteins
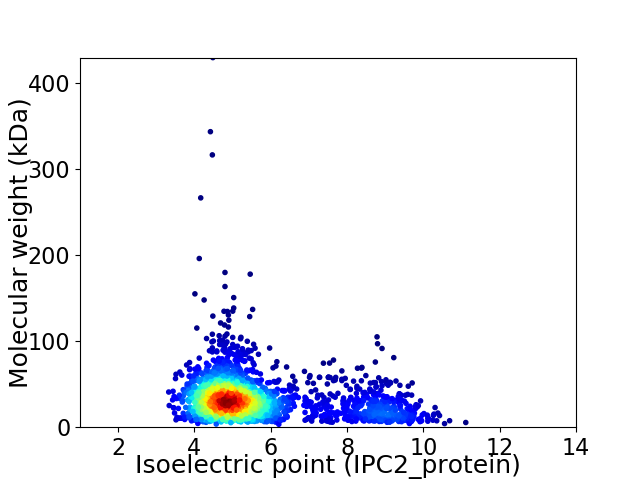
Virtual 2D-PAGE plot for 2209 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A347WJM2|A0A347WJM2_9LACT Uncharacterized protein OS=Suicoccus acidiformans OX=2036206 GN=CL176_04305 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.53KK3 pKa = 10.41LLLLSASMLALSSQGFAPSVHH24 pKa = 6.11AQNNADD30 pKa = 3.73VMAVTQTQQSQIQTDD45 pKa = 3.88YY46 pKa = 11.27DD47 pKa = 3.41QAIQSFQSQYY57 pKa = 10.6PNTHH61 pKa = 5.55ITDD64 pKa = 3.38VDD66 pKa = 3.27IHH68 pKa = 6.0LHH70 pKa = 5.09QEE72 pKa = 3.7TGHH75 pKa = 5.5FVVRR79 pKa = 11.84VLGADD84 pKa = 3.35QSQEE88 pKa = 4.11YY89 pKa = 10.07QLITYY94 pKa = 10.2NGEE97 pKa = 3.63QFNASRR103 pKa = 11.84PLEE106 pKa = 4.28EE107 pKa = 5.33ANSTSQTLNPAEE119 pKa = 4.27LASLEE124 pKa = 4.45TVTQNAIQQAQSDD137 pKa = 4.24KK138 pKa = 11.01AVNWSLEE145 pKa = 3.92GHH147 pKa = 6.85EE148 pKa = 4.96DD149 pKa = 3.25TDD151 pKa = 3.73APRR154 pKa = 11.84WDD156 pKa = 3.33VTFIEE161 pKa = 5.18DD162 pKa = 3.61NQEE165 pKa = 3.64KK166 pKa = 10.17NVAVDD171 pKa = 3.36ATTGEE176 pKa = 4.75VIDD179 pKa = 4.28STQDD183 pKa = 3.01SATQEE188 pKa = 4.27SADD191 pKa = 3.8TQQAPNASQNTDD203 pKa = 2.45QDD205 pKa = 3.56RR206 pKa = 11.84SDD208 pKa = 3.78NSDD211 pKa = 2.59ARR213 pKa = 11.84HH214 pKa = 6.77FYY216 pKa = 10.01RR217 pKa = 11.84QFYY220 pKa = 10.45SDD222 pKa = 3.8YY223 pKa = 9.96NGTRR227 pKa = 11.84PTVRR231 pKa = 11.84DD232 pKa = 2.96AGYY235 pKa = 10.36QIRR238 pKa = 11.84NTPSNSSQTTGPATSTVGQYY258 pKa = 11.37DD259 pKa = 3.73DD260 pKa = 6.13DD261 pKa = 5.1DD262 pKa = 3.85WDD264 pKa = 5.68DD265 pKa = 3.93YY266 pKa = 12.15DD267 pKa = 6.67DD268 pKa = 4.92YY269 pKa = 12.0DD270 pKa = 6.52DD271 pKa = 5.65YY272 pKa = 12.14DD273 pKa = 6.15DD274 pKa = 6.6YY275 pKa = 12.14DD276 pKa = 5.63DD277 pKa = 6.14DD278 pKa = 3.94WDD280 pKa = 6.21DD281 pKa = 3.97YY282 pKa = 12.14DD283 pKa = 6.05DD284 pKa = 4.65YY285 pKa = 12.01NDD287 pKa = 3.53YY288 pKa = 11.54DD289 pKa = 5.69DD290 pKa = 6.13YY291 pKa = 12.07DD292 pKa = 6.1GDD294 pKa = 4.67DD295 pKa = 3.78DD296 pKa = 4.58
MM1 pKa = 7.65KK2 pKa = 10.53KK3 pKa = 10.41LLLLSASMLALSSQGFAPSVHH24 pKa = 6.11AQNNADD30 pKa = 3.73VMAVTQTQQSQIQTDD45 pKa = 3.88YY46 pKa = 11.27DD47 pKa = 3.41QAIQSFQSQYY57 pKa = 10.6PNTHH61 pKa = 5.55ITDD64 pKa = 3.38VDD66 pKa = 3.27IHH68 pKa = 6.0LHH70 pKa = 5.09QEE72 pKa = 3.7TGHH75 pKa = 5.5FVVRR79 pKa = 11.84VLGADD84 pKa = 3.35QSQEE88 pKa = 4.11YY89 pKa = 10.07QLITYY94 pKa = 10.2NGEE97 pKa = 3.63QFNASRR103 pKa = 11.84PLEE106 pKa = 4.28EE107 pKa = 5.33ANSTSQTLNPAEE119 pKa = 4.27LASLEE124 pKa = 4.45TVTQNAIQQAQSDD137 pKa = 4.24KK138 pKa = 11.01AVNWSLEE145 pKa = 3.92GHH147 pKa = 6.85EE148 pKa = 4.96DD149 pKa = 3.25TDD151 pKa = 3.73APRR154 pKa = 11.84WDD156 pKa = 3.33VTFIEE161 pKa = 5.18DD162 pKa = 3.61NQEE165 pKa = 3.64KK166 pKa = 10.17NVAVDD171 pKa = 3.36ATTGEE176 pKa = 4.75VIDD179 pKa = 4.28STQDD183 pKa = 3.01SATQEE188 pKa = 4.27SADD191 pKa = 3.8TQQAPNASQNTDD203 pKa = 2.45QDD205 pKa = 3.56RR206 pKa = 11.84SDD208 pKa = 3.78NSDD211 pKa = 2.59ARR213 pKa = 11.84HH214 pKa = 6.77FYY216 pKa = 10.01RR217 pKa = 11.84QFYY220 pKa = 10.45SDD222 pKa = 3.8YY223 pKa = 9.96NGTRR227 pKa = 11.84PTVRR231 pKa = 11.84DD232 pKa = 2.96AGYY235 pKa = 10.36QIRR238 pKa = 11.84NTPSNSSQTTGPATSTVGQYY258 pKa = 11.37DD259 pKa = 3.73DD260 pKa = 6.13DD261 pKa = 5.1DD262 pKa = 3.85WDD264 pKa = 5.68DD265 pKa = 3.93YY266 pKa = 12.15DD267 pKa = 6.67DD268 pKa = 4.92YY269 pKa = 12.0DD270 pKa = 6.52DD271 pKa = 5.65YY272 pKa = 12.14DD273 pKa = 6.15DD274 pKa = 6.6YY275 pKa = 12.14DD276 pKa = 5.63DD277 pKa = 6.14DD278 pKa = 3.94WDD280 pKa = 6.21DD281 pKa = 3.97YY282 pKa = 12.14DD283 pKa = 6.05DD284 pKa = 4.65YY285 pKa = 12.01NDD287 pKa = 3.53YY288 pKa = 11.54DD289 pKa = 5.69DD290 pKa = 6.13YY291 pKa = 12.07DD292 pKa = 6.1GDD294 pKa = 4.67DD295 pKa = 3.78DD296 pKa = 4.58
Molecular weight: 33.34 kDa
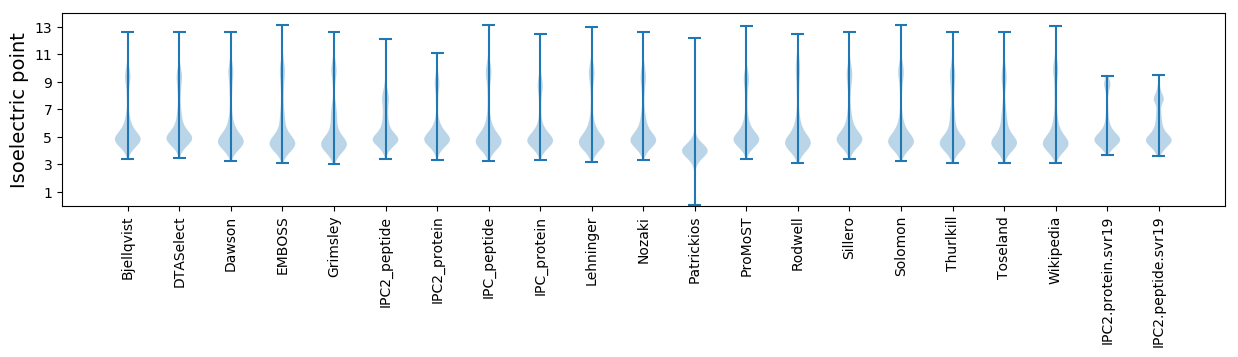
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A347WHK8|A0A347WHK8_9LACT Guanine permease OS=Suicoccus acidiformans OX=2036206 GN=CL176_00155 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 8.89KK9 pKa = 7.7RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 7.95KK14 pKa = 8.03VHH16 pKa = 5.72GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.14NGRR28 pKa = 11.84NVLKK32 pKa = 10.51RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.59GRR39 pKa = 11.84KK40 pKa = 8.75KK41 pKa = 10.39LASS44 pKa = 3.43
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 8.89KK9 pKa = 7.7RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 7.95KK14 pKa = 8.03VHH16 pKa = 5.72GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.14NGRR28 pKa = 11.84NVLKK32 pKa = 10.51RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.59GRR39 pKa = 11.84KK40 pKa = 8.75KK41 pKa = 10.39LASS44 pKa = 3.43
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
678505 |
28 |
3877 |
307.2 |
34.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.106 ± 0.057 | 0.508 ± 0.013 |
5.956 ± 0.047 | 7.785 ± 0.06 |
3.978 ± 0.034 | 6.637 ± 0.05 |
2.097 ± 0.025 | 7.225 ± 0.052 |
4.97 ± 0.05 | 9.656 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.601 ± 0.031 | 4.359 ± 0.038 |
3.646 ± 0.053 | 4.821 ± 0.047 |
4.449 ± 0.042 | 5.785 ± 0.038 |
5.546 ± 0.054 | 6.992 ± 0.044 |
0.876 ± 0.017 | 4.007 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
