
Pseudokineococcus lusitanus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Kineosporiales; Kineosporiaceae; Pseudokineococcus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
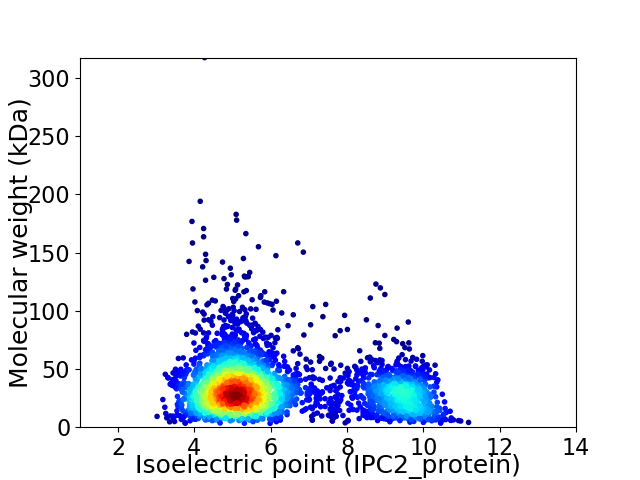
Virtual 2D-PAGE plot for 3389 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N1G9Z6|A0A3N1G9Z6_9ACTN MsuE subfamily FMN reductase OS=Pseudokineococcus lusitanus OX=763993 GN=EDC03_2990 PE=4 SV=1
MM1 pKa = 7.33TRR3 pKa = 11.84TALTSTPRR11 pKa = 11.84RR12 pKa = 11.84THH14 pKa = 5.53ARR16 pKa = 11.84RR17 pKa = 11.84AASAVVALTTAAALAACGGGSDD39 pKa = 4.34GGNAGGDD46 pKa = 3.9EE47 pKa = 4.16NTLTWWHH54 pKa = 6.39NSNTDD59 pKa = 3.26PGAGYY64 pKa = 10.26YY65 pKa = 10.37EE66 pKa = 4.13EE67 pKa = 4.1VAAAFEE73 pKa = 4.25EE74 pKa = 4.68ANPGTTIEE82 pKa = 4.27ISALPHH88 pKa = 6.21EE89 pKa = 5.45DD90 pKa = 2.74MLTRR94 pKa = 11.84LNAAFQSGDD103 pKa = 3.43VPDD106 pKa = 4.19VYY108 pKa = 10.08MEE110 pKa = 4.22RR111 pKa = 11.84GGGEE115 pKa = 3.91LADD118 pKa = 3.74HH119 pKa = 6.42VEE121 pKa = 4.54AGLTMDD127 pKa = 4.74LSEE130 pKa = 4.72GAADD134 pKa = 4.08TIEE137 pKa = 4.66KK138 pKa = 10.23IGGSVAGWQVDD149 pKa = 3.47GATYY153 pKa = 10.84ALPFSVGVVGFWYY166 pKa = 8.55NTEE169 pKa = 4.18LFEE172 pKa = 4.79QAGITEE178 pKa = 4.87APTTMEE184 pKa = 4.26EE185 pKa = 4.16LDD187 pKa = 3.94AAVQALKK194 pKa = 10.76DD195 pKa = 3.7AGIQPISVGAGDD207 pKa = 3.99TWPAAHH213 pKa = 6.13YY214 pKa = 8.78WYY216 pKa = 10.53YY217 pKa = 10.88LAVRR221 pKa = 11.84EE222 pKa = 4.44CGQDD226 pKa = 3.36VLTDD230 pKa = 3.7AVTSLDD236 pKa = 3.74FSDD239 pKa = 3.69EE240 pKa = 4.3CFVRR244 pKa = 11.84AGEE247 pKa = 4.22DD248 pKa = 3.19LEE250 pKa = 5.37AFIGTEE256 pKa = 3.79PFNEE260 pKa = 4.4GFLATPAQVGPASASGLLASEE281 pKa = 4.14QVAMEE286 pKa = 4.54LAGHH290 pKa = 6.32WEE292 pKa = 4.01PGVMQGLTEE301 pKa = 4.36GGEE304 pKa = 4.34GLGEE308 pKa = 3.99KK309 pKa = 9.37TGWFAFPTVEE319 pKa = 4.3GGEE322 pKa = 4.35GAPDD326 pKa = 3.45AALGGGDD333 pKa = 3.7AWACAADD340 pKa = 4.12APPVCVDD347 pKa = 3.62FVNYY351 pKa = 10.22LLSDD355 pKa = 4.07EE356 pKa = 4.52VQTGFAEE363 pKa = 4.26RR364 pKa = 11.84DD365 pKa = 3.13MGLPTNPAATGAVSDD380 pKa = 4.55PALADD385 pKa = 4.62LIAVRR390 pKa = 11.84DD391 pKa = 3.82SAPYY395 pKa = 8.02VQLYY399 pKa = 10.15FDD401 pKa = 3.63TAFGEE406 pKa = 4.49NVGGAMNDD414 pKa = 3.81AIALMFAGQGSAQDD428 pKa = 3.76VVDD431 pKa = 4.22ATQSAAGTAA440 pKa = 3.53
MM1 pKa = 7.33TRR3 pKa = 11.84TALTSTPRR11 pKa = 11.84RR12 pKa = 11.84THH14 pKa = 5.53ARR16 pKa = 11.84RR17 pKa = 11.84AASAVVALTTAAALAACGGGSDD39 pKa = 4.34GGNAGGDD46 pKa = 3.9EE47 pKa = 4.16NTLTWWHH54 pKa = 6.39NSNTDD59 pKa = 3.26PGAGYY64 pKa = 10.26YY65 pKa = 10.37EE66 pKa = 4.13EE67 pKa = 4.1VAAAFEE73 pKa = 4.25EE74 pKa = 4.68ANPGTTIEE82 pKa = 4.27ISALPHH88 pKa = 6.21EE89 pKa = 5.45DD90 pKa = 2.74MLTRR94 pKa = 11.84LNAAFQSGDD103 pKa = 3.43VPDD106 pKa = 4.19VYY108 pKa = 10.08MEE110 pKa = 4.22RR111 pKa = 11.84GGGEE115 pKa = 3.91LADD118 pKa = 3.74HH119 pKa = 6.42VEE121 pKa = 4.54AGLTMDD127 pKa = 4.74LSEE130 pKa = 4.72GAADD134 pKa = 4.08TIEE137 pKa = 4.66KK138 pKa = 10.23IGGSVAGWQVDD149 pKa = 3.47GATYY153 pKa = 10.84ALPFSVGVVGFWYY166 pKa = 8.55NTEE169 pKa = 4.18LFEE172 pKa = 4.79QAGITEE178 pKa = 4.87APTTMEE184 pKa = 4.26EE185 pKa = 4.16LDD187 pKa = 3.94AAVQALKK194 pKa = 10.76DD195 pKa = 3.7AGIQPISVGAGDD207 pKa = 3.99TWPAAHH213 pKa = 6.13YY214 pKa = 8.78WYY216 pKa = 10.53YY217 pKa = 10.88LAVRR221 pKa = 11.84EE222 pKa = 4.44CGQDD226 pKa = 3.36VLTDD230 pKa = 3.7AVTSLDD236 pKa = 3.74FSDD239 pKa = 3.69EE240 pKa = 4.3CFVRR244 pKa = 11.84AGEE247 pKa = 4.22DD248 pKa = 3.19LEE250 pKa = 5.37AFIGTEE256 pKa = 3.79PFNEE260 pKa = 4.4GFLATPAQVGPASASGLLASEE281 pKa = 4.14QVAMEE286 pKa = 4.54LAGHH290 pKa = 6.32WEE292 pKa = 4.01PGVMQGLTEE301 pKa = 4.36GGEE304 pKa = 4.34GLGEE308 pKa = 3.99KK309 pKa = 9.37TGWFAFPTVEE319 pKa = 4.3GGEE322 pKa = 4.35GAPDD326 pKa = 3.45AALGGGDD333 pKa = 3.7AWACAADD340 pKa = 4.12APPVCVDD347 pKa = 3.62FVNYY351 pKa = 10.22LLSDD355 pKa = 4.07EE356 pKa = 4.52VQTGFAEE363 pKa = 4.26RR364 pKa = 11.84DD365 pKa = 3.13MGLPTNPAATGAVSDD380 pKa = 4.55PALADD385 pKa = 4.62LIAVRR390 pKa = 11.84DD391 pKa = 3.82SAPYY395 pKa = 8.02VQLYY399 pKa = 10.15FDD401 pKa = 3.63TAFGEE406 pKa = 4.49NVGGAMNDD414 pKa = 3.81AIALMFAGQGSAQDD428 pKa = 3.76VVDD431 pKa = 4.22ATQSAAGTAA440 pKa = 3.53
Molecular weight: 45.31 kDa
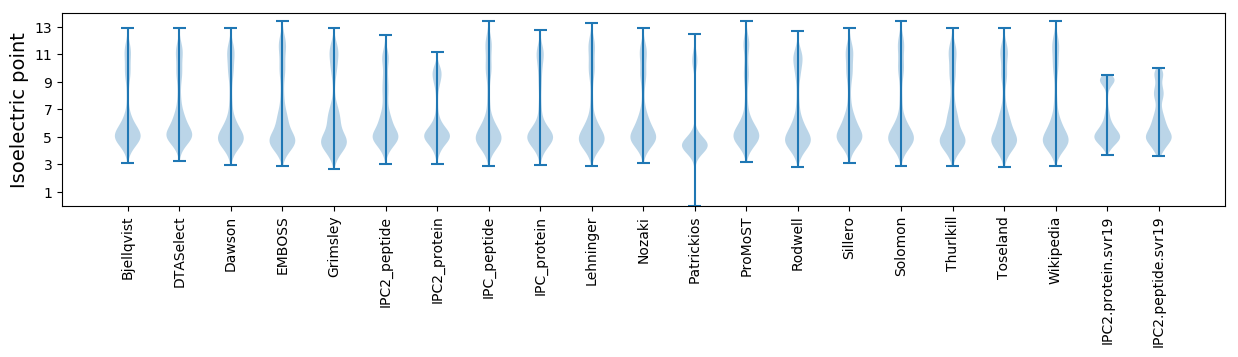
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N1HMB5|A0A3N1HMB5_9ACTN tRNA pseudouridine synthase B OS=Pseudokineococcus lusitanus OX=763993 GN=truB PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.64HH17 pKa = 5.77RR18 pKa = 11.84KK19 pKa = 7.85LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84HH26 pKa = 3.6QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.64HH17 pKa = 5.77RR18 pKa = 11.84KK19 pKa = 7.85LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84HH26 pKa = 3.6QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1148759 |
29 |
3317 |
339.0 |
35.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.619 ± 0.072 | 0.567 ± 0.009 |
6.622 ± 0.04 | 5.314 ± 0.038 |
2.003 ± 0.022 | 10.391 ± 0.041 |
1.906 ± 0.022 | 1.532 ± 0.027 |
1.01 ± 0.018 | 10.746 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.312 ± 0.015 | 1.048 ± 0.018 |
6.592 ± 0.036 | 2.301 ± 0.022 |
8.509 ± 0.05 | 4.457 ± 0.028 |
6.004 ± 0.046 | 11.154 ± 0.045 |
1.39 ± 0.016 | 1.524 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
