
Vitis riparia x Vitis cinerea
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; rosids incertae sedis; Vitales; Vitaceae; Viteae; Vitis
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
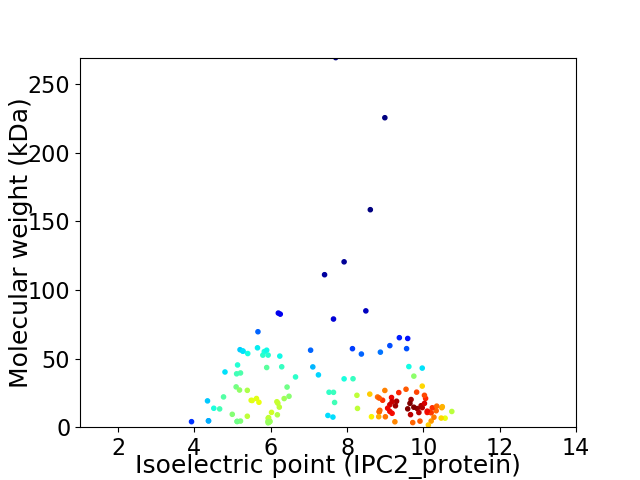
Virtual 2D-PAGE plot for 138 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
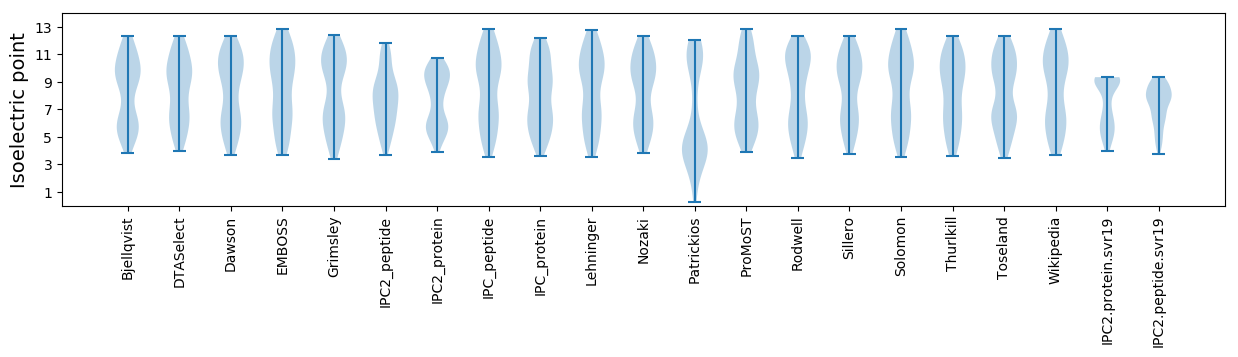
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A654IBI0|A0A654IBI0_9ROSI 50S ribosomal protein L33 chloroplastic OS=Vitis riparia x Vitis cinerea OX=2018007 GN=rpl33 PE=3 SV=1
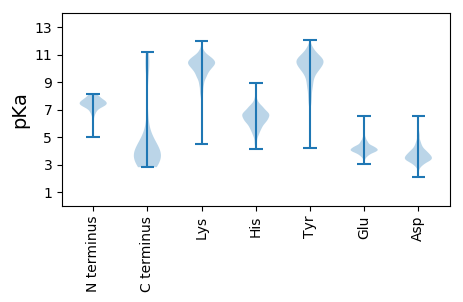
MM1 pKa = 6.86EE2 pKa = 4.58TATLVAISISGLLVSLTGYY21 pKa = 11.01ALYY24 pKa = 9.16TAFGQPSQQLRR35 pKa = 11.84DD36 pKa = 3.65PFEE39 pKa = 4.0EE40 pKa = 5.39HH41 pKa = 7.25GDD43 pKa = 3.49
MM1 pKa = 6.86EE2 pKa = 4.58TATLVAISISGLLVSLTGYY21 pKa = 11.01ALYY24 pKa = 9.16TAFGQPSQQLRR35 pKa = 11.84DD36 pKa = 3.65PFEE39 pKa = 4.0EE40 pKa = 5.39HH41 pKa = 7.25GDD43 pKa = 3.49
Molecular weight: 4.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A654IBM9|A0A654IBM9_9ROSI Rps19 protein OS=Vitis riparia x Vitis cinerea OX=2018007 GN=rps19 PE=3 SV=1
MM1 pKa = 6.88VQLHH5 pKa = 6.19NSFFFITSMFVPRR18 pKa = 11.84GTASPALWKK27 pKa = 9.93WFVSRR32 pKa = 11.84EE33 pKa = 4.05IPTGAPSSNGTIIPIPIPSFPLLVYY58 pKa = 9.56LHH60 pKa = 6.06SRR62 pKa = 11.84KK63 pKa = 9.61FIRR66 pKa = 11.84FIDD69 pKa = 3.8RR70 pKa = 11.84AKK72 pKa = 10.84SGVLVRR78 pKa = 11.84ASHH81 pKa = 6.95PIILLSDD88 pKa = 4.14IIRR91 pKa = 11.84ISSSEE96 pKa = 4.06TIARR100 pKa = 11.84RR101 pKa = 11.84ASFRR105 pKa = 11.84FVPVLHH111 pKa = 6.6FLLLEE116 pKa = 4.31SKK118 pKa = 11.15GDD120 pKa = 3.44ISYY123 pKa = 11.02LEE125 pKa = 4.3SFCGVLRR132 pKa = 11.84LLFFRR137 pKa = 11.84TLLSLPRR144 pKa = 11.84DD145 pKa = 3.24RR146 pKa = 11.84SAKK149 pKa = 9.65RR150 pKa = 11.84EE151 pKa = 3.56RR152 pKa = 11.84ARR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84KK157 pKa = 9.5GQALRR162 pKa = 11.84PNGNEE167 pKa = 3.39QRR169 pKa = 11.84RR170 pKa = 11.84NDD172 pKa = 3.46KK173 pKa = 9.49MRR175 pKa = 11.84CPGHH179 pKa = 5.89PHH181 pKa = 6.52LEE183 pKa = 3.89RR184 pKa = 11.84RR185 pKa = 11.84VEE187 pKa = 4.14GFGPVAFPVPPSSGGACVGGVPPEE211 pKa = 4.18IGLEE215 pKa = 3.92ALALPTSRR223 pKa = 11.84QLRR226 pKa = 11.84AVGDD230 pKa = 3.89DD231 pKa = 4.18YY232 pKa = 11.59YY233 pKa = 11.63QKK235 pKa = 11.53APMKK239 pKa = 10.14MNISHH244 pKa = 6.86GGVCICMLGVLLSKK258 pKa = 9.33TKK260 pKa = 10.38KK261 pKa = 9.48IQFTQRR267 pKa = 11.84LPLGSEE273 pKa = 3.44LHH275 pKa = 5.92MGKK278 pKa = 9.29EE279 pKa = 4.39RR280 pKa = 11.84CCLRR284 pKa = 11.84GIDD287 pKa = 4.28HH288 pKa = 6.82LHH290 pKa = 6.57GPTSHH295 pKa = 6.86SICGNFMIYY304 pKa = 9.85KK305 pKa = 10.33RR306 pKa = 11.84SLTTDD311 pKa = 2.68RR312 pKa = 11.84LMFEE316 pKa = 4.38HH317 pKa = 7.53DD318 pKa = 3.13EE319 pKa = 4.18SLRR322 pKa = 11.84ADD324 pKa = 4.54LLPIKK329 pKa = 9.93NFPASYY335 pKa = 10.85EE336 pKa = 4.04NGKK339 pKa = 9.84LKK341 pKa = 10.73HH342 pKa = 6.17FLHH345 pKa = 6.92RR346 pKa = 11.84WMKK349 pKa = 10.66NRR351 pKa = 11.84IKK353 pKa = 11.09NNFWFTMFPEE363 pKa = 3.77KK364 pKa = 10.34RR365 pKa = 11.84YY366 pKa = 9.81FRR368 pKa = 11.84EE369 pKa = 4.23TTSTTEE375 pKa = 4.08VAIHH379 pKa = 6.02TNPLTDD385 pKa = 4.25LYY387 pKa = 11.56APIGTGNSRR396 pKa = 11.84TSGWYY401 pKa = 4.21TTIMKK406 pKa = 10.37LPFIFFIRR414 pKa = 11.84IGFMMASSGGSRR426 pKa = 11.84SLLRR430 pKa = 11.84QLQKK434 pKa = 11.34DD435 pKa = 3.29KK436 pKa = 11.26LRR438 pKa = 11.84WNRR441 pKa = 11.84EE442 pKa = 3.64SSVEE446 pKa = 4.14FIIAINARR454 pKa = 11.84SQSSGCDD461 pKa = 3.19ASRR464 pKa = 11.84HH465 pKa = 5.07FFDD468 pKa = 5.09GPRR471 pKa = 11.84PPGLPARR478 pKa = 11.84SEE480 pKa = 4.1EE481 pKa = 4.13FMGRR485 pKa = 11.84QAGEE489 pKa = 4.29TEE491 pKa = 3.96WAGTTNQAKK500 pKa = 9.82PSSRR504 pKa = 11.84RR505 pKa = 3.39
MM1 pKa = 6.88VQLHH5 pKa = 6.19NSFFFITSMFVPRR18 pKa = 11.84GTASPALWKK27 pKa = 9.93WFVSRR32 pKa = 11.84EE33 pKa = 4.05IPTGAPSSNGTIIPIPIPSFPLLVYY58 pKa = 9.56LHH60 pKa = 6.06SRR62 pKa = 11.84KK63 pKa = 9.61FIRR66 pKa = 11.84FIDD69 pKa = 3.8RR70 pKa = 11.84AKK72 pKa = 10.84SGVLVRR78 pKa = 11.84ASHH81 pKa = 6.95PIILLSDD88 pKa = 4.14IIRR91 pKa = 11.84ISSSEE96 pKa = 4.06TIARR100 pKa = 11.84RR101 pKa = 11.84ASFRR105 pKa = 11.84FVPVLHH111 pKa = 6.6FLLLEE116 pKa = 4.31SKK118 pKa = 11.15GDD120 pKa = 3.44ISYY123 pKa = 11.02LEE125 pKa = 4.3SFCGVLRR132 pKa = 11.84LLFFRR137 pKa = 11.84TLLSLPRR144 pKa = 11.84DD145 pKa = 3.24RR146 pKa = 11.84SAKK149 pKa = 9.65RR150 pKa = 11.84EE151 pKa = 3.56RR152 pKa = 11.84ARR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84KK157 pKa = 9.5GQALRR162 pKa = 11.84PNGNEE167 pKa = 3.39QRR169 pKa = 11.84RR170 pKa = 11.84NDD172 pKa = 3.46KK173 pKa = 9.49MRR175 pKa = 11.84CPGHH179 pKa = 5.89PHH181 pKa = 6.52LEE183 pKa = 3.89RR184 pKa = 11.84RR185 pKa = 11.84VEE187 pKa = 4.14GFGPVAFPVPPSSGGACVGGVPPEE211 pKa = 4.18IGLEE215 pKa = 3.92ALALPTSRR223 pKa = 11.84QLRR226 pKa = 11.84AVGDD230 pKa = 3.89DD231 pKa = 4.18YY232 pKa = 11.59YY233 pKa = 11.63QKK235 pKa = 11.53APMKK239 pKa = 10.14MNISHH244 pKa = 6.86GGVCICMLGVLLSKK258 pKa = 9.33TKK260 pKa = 10.38KK261 pKa = 9.48IQFTQRR267 pKa = 11.84LPLGSEE273 pKa = 3.44LHH275 pKa = 5.92MGKK278 pKa = 9.29EE279 pKa = 4.39RR280 pKa = 11.84CCLRR284 pKa = 11.84GIDD287 pKa = 4.28HH288 pKa = 6.82LHH290 pKa = 6.57GPTSHH295 pKa = 6.86SICGNFMIYY304 pKa = 9.85KK305 pKa = 10.33RR306 pKa = 11.84SLTTDD311 pKa = 2.68RR312 pKa = 11.84LMFEE316 pKa = 4.38HH317 pKa = 7.53DD318 pKa = 3.13EE319 pKa = 4.18SLRR322 pKa = 11.84ADD324 pKa = 4.54LLPIKK329 pKa = 9.93NFPASYY335 pKa = 10.85EE336 pKa = 4.04NGKK339 pKa = 9.84LKK341 pKa = 10.73HH342 pKa = 6.17FLHH345 pKa = 6.92RR346 pKa = 11.84WMKK349 pKa = 10.66NRR351 pKa = 11.84IKK353 pKa = 11.09NNFWFTMFPEE363 pKa = 3.77KK364 pKa = 10.34RR365 pKa = 11.84YY366 pKa = 9.81FRR368 pKa = 11.84EE369 pKa = 4.23TTSTTEE375 pKa = 4.08VAIHH379 pKa = 6.02TNPLTDD385 pKa = 4.25LYY387 pKa = 11.56APIGTGNSRR396 pKa = 11.84TSGWYY401 pKa = 4.21TTIMKK406 pKa = 10.37LPFIFFIRR414 pKa = 11.84IGFMMASSGGSRR426 pKa = 11.84SLLRR430 pKa = 11.84QLQKK434 pKa = 11.34DD435 pKa = 3.29KK436 pKa = 11.26LRR438 pKa = 11.84WNRR441 pKa = 11.84EE442 pKa = 3.64SSVEE446 pKa = 4.14FIIAINARR454 pKa = 11.84SQSSGCDD461 pKa = 3.19ASRR464 pKa = 11.84HH465 pKa = 5.07FFDD468 pKa = 5.09GPRR471 pKa = 11.84PPGLPARR478 pKa = 11.84SEE480 pKa = 4.1EE481 pKa = 4.13FMGRR485 pKa = 11.84QAGEE489 pKa = 4.29TEE491 pKa = 3.96WAGTTNQAKK500 pKa = 9.82PSSRR504 pKa = 11.84RR505 pKa = 3.39
Molecular weight: 57.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
37120 |
13 |
2300 |
269.0 |
30.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.884 ± 0.43 | 1.272 ± 0.081 |
3.858 ± 0.192 | 5.0 ± 0.236 |
5.595 ± 0.238 | 6.915 ± 0.393 |
2.435 ± 0.14 | 8.314 ± 0.242 |
4.925 ± 0.386 | 10.291 ± 0.228 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.465 ± 0.077 | 4.34 ± 0.291 |
4.733 ± 0.221 | 3.44 ± 0.143 |
6.447 ± 0.268 | 7.977 ± 0.286 |
5.21 ± 0.141 | 5.722 ± 0.266 |
1.619 ± 0.124 | 3.559 ± 0.139 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
