
Menangle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Pararubulavirus; Menangle pararubulavirus
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
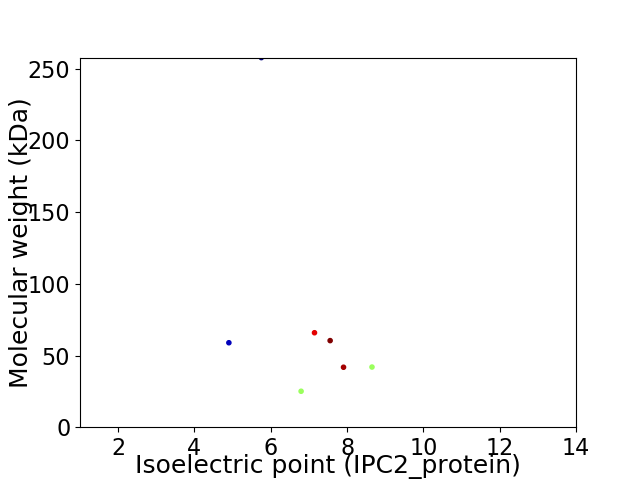
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q91MK2|Q91MK2_9MONO Isoform of Q91MK1 Non-structural protein V OS=Menangle virus OX=152219 GN=V/P PE=3 SV=1
MM1 pKa = 7.4SSVFRR6 pKa = 11.84AFEE9 pKa = 4.3LFTLEE14 pKa = 4.59QEE16 pKa = 4.52QNEE19 pKa = 4.39HH20 pKa = 6.46GNDD23 pKa = 3.11IEE25 pKa = 5.19LPPEE29 pKa = 3.96TLRR32 pKa = 11.84TNIKK36 pKa = 10.08VCTLNNQEE44 pKa = 3.8PQARR48 pKa = 11.84HH49 pKa = 6.55DD50 pKa = 3.8MMCFCLRR57 pKa = 11.84LIASNSARR65 pKa = 11.84AAHH68 pKa = 5.96KK69 pKa = 9.34TGAILTLLSLPTAMMQNHH87 pKa = 6.33LRR89 pKa = 11.84IADD92 pKa = 4.31RR93 pKa = 11.84SPDD96 pKa = 3.38ADD98 pKa = 3.45IEE100 pKa = 4.11RR101 pKa = 11.84LEE103 pKa = 4.42IDD105 pKa = 3.46GFEE108 pKa = 4.53PGTFRR113 pKa = 11.84LRR115 pKa = 11.84ANARR119 pKa = 11.84TPMTNGEE126 pKa = 4.12VTALNLMAQDD136 pKa = 5.46LPDD139 pKa = 3.9TYY141 pKa = 11.76SNDD144 pKa = 3.75TPFLNPNTEE153 pKa = 4.16TEE155 pKa = 4.06QCDD158 pKa = 3.82EE159 pKa = 4.3MEE161 pKa = 4.17QFLNAIYY168 pKa = 10.51SVLVQVWVTVCKK180 pKa = 10.65CMTAHH185 pKa = 6.77DD186 pKa = 4.33QPTGSDD192 pKa = 3.06EE193 pKa = 4.13RR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.17YY199 pKa = 8.02QQQGRR204 pKa = 11.84LDD206 pKa = 3.26QRR208 pKa = 11.84YY209 pKa = 8.96ALQPEE214 pKa = 4.98LRR216 pKa = 11.84RR217 pKa = 11.84QIQTCIRR224 pKa = 11.84SSLTIRR230 pKa = 11.84QFLTHH235 pKa = 6.73EE236 pKa = 4.26LQTARR241 pKa = 11.84KK242 pKa = 8.65QGAITGKK249 pKa = 9.08YY250 pKa = 7.82YY251 pKa = 11.55AMVGDD256 pKa = 3.45IGKK259 pKa = 10.43YY260 pKa = 9.05IDD262 pKa = 3.75NAGMSAFFMTMRR274 pKa = 11.84FALGTKK280 pKa = 8.92WPPLALAAFSGEE292 pKa = 3.98LLKK295 pKa = 11.09LKK297 pKa = 10.68SLMQLYY303 pKa = 10.2RR304 pKa = 11.84SLGDD308 pKa = 3.15RR309 pKa = 11.84ARR311 pKa = 11.84YY312 pKa = 7.24MALLEE317 pKa = 4.08MSEE320 pKa = 4.07MMEE323 pKa = 4.3FAPANYY329 pKa = 8.35PLCYY333 pKa = 9.75SYY335 pKa = 11.89AMGIGSVQDD344 pKa = 3.3PMMRR348 pKa = 11.84NYY350 pKa = 9.81TFARR354 pKa = 11.84PFLNPAYY361 pKa = 9.55FQLGVEE367 pKa = 4.29TANRR371 pKa = 11.84QQGSVDD377 pKa = 2.93KK378 pKa = 11.47AMAAEE383 pKa = 4.62LGLTEE388 pKa = 4.82DD389 pKa = 4.34EE390 pKa = 4.57KK391 pKa = 11.47RR392 pKa = 11.84DD393 pKa = 3.28MSAAVTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GGNQAQEE412 pKa = 4.86LINVMGARR420 pKa = 11.84QGRR423 pKa = 11.84DD424 pKa = 2.79QGRR427 pKa = 11.84RR428 pKa = 11.84GNRR431 pKa = 11.84DD432 pKa = 2.9YY433 pKa = 11.69DD434 pKa = 3.7VVEE437 pKa = 4.34EE438 pKa = 4.18NEE440 pKa = 4.23EE441 pKa = 4.29TEE443 pKa = 4.12SDD445 pKa = 3.49SDD447 pKa = 3.61NDD449 pKa = 3.68EE450 pKa = 3.96EE451 pKa = 5.56QEE453 pKa = 4.07IQNRR457 pKa = 11.84PLPPIPQMPQNIDD470 pKa = 3.02WEE472 pKa = 4.5VRR474 pKa = 11.84LAEE477 pKa = 3.97IEE479 pKa = 4.2RR480 pKa = 11.84RR481 pKa = 11.84NQQMAARR488 pKa = 11.84DD489 pKa = 3.64RR490 pKa = 11.84PQAVVTADD498 pKa = 3.4VHH500 pKa = 6.63QEE502 pKa = 3.71PADD505 pKa = 3.74ARR507 pKa = 11.84VDD509 pKa = 3.88EE510 pKa = 4.44QDD512 pKa = 3.34MLLDD516 pKa = 4.26LDD518 pKa = 3.95MM519 pKa = 6.44
MM1 pKa = 7.4SSVFRR6 pKa = 11.84AFEE9 pKa = 4.3LFTLEE14 pKa = 4.59QEE16 pKa = 4.52QNEE19 pKa = 4.39HH20 pKa = 6.46GNDD23 pKa = 3.11IEE25 pKa = 5.19LPPEE29 pKa = 3.96TLRR32 pKa = 11.84TNIKK36 pKa = 10.08VCTLNNQEE44 pKa = 3.8PQARR48 pKa = 11.84HH49 pKa = 6.55DD50 pKa = 3.8MMCFCLRR57 pKa = 11.84LIASNSARR65 pKa = 11.84AAHH68 pKa = 5.96KK69 pKa = 9.34TGAILTLLSLPTAMMQNHH87 pKa = 6.33LRR89 pKa = 11.84IADD92 pKa = 4.31RR93 pKa = 11.84SPDD96 pKa = 3.38ADD98 pKa = 3.45IEE100 pKa = 4.11RR101 pKa = 11.84LEE103 pKa = 4.42IDD105 pKa = 3.46GFEE108 pKa = 4.53PGTFRR113 pKa = 11.84LRR115 pKa = 11.84ANARR119 pKa = 11.84TPMTNGEE126 pKa = 4.12VTALNLMAQDD136 pKa = 5.46LPDD139 pKa = 3.9TYY141 pKa = 11.76SNDD144 pKa = 3.75TPFLNPNTEE153 pKa = 4.16TEE155 pKa = 4.06QCDD158 pKa = 3.82EE159 pKa = 4.3MEE161 pKa = 4.17QFLNAIYY168 pKa = 10.51SVLVQVWVTVCKK180 pKa = 10.65CMTAHH185 pKa = 6.77DD186 pKa = 4.33QPTGSDD192 pKa = 3.06EE193 pKa = 4.13RR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.17YY199 pKa = 8.02QQQGRR204 pKa = 11.84LDD206 pKa = 3.26QRR208 pKa = 11.84YY209 pKa = 8.96ALQPEE214 pKa = 4.98LRR216 pKa = 11.84RR217 pKa = 11.84QIQTCIRR224 pKa = 11.84SSLTIRR230 pKa = 11.84QFLTHH235 pKa = 6.73EE236 pKa = 4.26LQTARR241 pKa = 11.84KK242 pKa = 8.65QGAITGKK249 pKa = 9.08YY250 pKa = 7.82YY251 pKa = 11.55AMVGDD256 pKa = 3.45IGKK259 pKa = 10.43YY260 pKa = 9.05IDD262 pKa = 3.75NAGMSAFFMTMRR274 pKa = 11.84FALGTKK280 pKa = 8.92WPPLALAAFSGEE292 pKa = 3.98LLKK295 pKa = 11.09LKK297 pKa = 10.68SLMQLYY303 pKa = 10.2RR304 pKa = 11.84SLGDD308 pKa = 3.15RR309 pKa = 11.84ARR311 pKa = 11.84YY312 pKa = 7.24MALLEE317 pKa = 4.08MSEE320 pKa = 4.07MMEE323 pKa = 4.3FAPANYY329 pKa = 8.35PLCYY333 pKa = 9.75SYY335 pKa = 11.89AMGIGSVQDD344 pKa = 3.3PMMRR348 pKa = 11.84NYY350 pKa = 9.81TFARR354 pKa = 11.84PFLNPAYY361 pKa = 9.55FQLGVEE367 pKa = 4.29TANRR371 pKa = 11.84QQGSVDD377 pKa = 2.93KK378 pKa = 11.47AMAAEE383 pKa = 4.62LGLTEE388 pKa = 4.82DD389 pKa = 4.34EE390 pKa = 4.57KK391 pKa = 11.47RR392 pKa = 11.84DD393 pKa = 3.28MSAAVTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GGNQAQEE412 pKa = 4.86LINVMGARR420 pKa = 11.84QGRR423 pKa = 11.84DD424 pKa = 2.79QGRR427 pKa = 11.84RR428 pKa = 11.84GNRR431 pKa = 11.84DD432 pKa = 2.9YY433 pKa = 11.69DD434 pKa = 3.7VVEE437 pKa = 4.34EE438 pKa = 4.18NEE440 pKa = 4.23EE441 pKa = 4.29TEE443 pKa = 4.12SDD445 pKa = 3.49SDD447 pKa = 3.61NDD449 pKa = 3.68EE450 pKa = 3.96EE451 pKa = 5.56QEE453 pKa = 4.07IQNRR457 pKa = 11.84PLPPIPQMPQNIDD470 pKa = 3.02WEE472 pKa = 4.5VRR474 pKa = 11.84LAEE477 pKa = 3.97IEE479 pKa = 4.2RR480 pKa = 11.84RR481 pKa = 11.84NQQMAARR488 pKa = 11.84DD489 pKa = 3.64RR490 pKa = 11.84PQAVVTADD498 pKa = 3.4VHH500 pKa = 6.63QEE502 pKa = 3.71PADD505 pKa = 3.74ARR507 pKa = 11.84VDD509 pKa = 3.88EE510 pKa = 4.44QDD512 pKa = 3.34MLLDD516 pKa = 4.26LDD518 pKa = 3.95MM519 pKa = 6.44
Molecular weight: 58.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91MK1|Q91MK1_9MONO Phosphoprotein OS=Menangle virus OX=152219 GN=V/P PE=1 SV=1
MM1 pKa = 8.01ALRR4 pKa = 11.84QATIPVDD11 pKa = 3.38VDD13 pKa = 3.34KK14 pKa = 11.42DD15 pKa = 3.94SEE17 pKa = 4.5KK18 pKa = 11.38NNLNPFPIIPVTRR31 pKa = 11.84DD32 pKa = 3.25DD33 pKa = 4.13GSSTGRR39 pKa = 11.84LVKK42 pKa = 9.77QLRR45 pKa = 11.84VKK47 pKa = 10.76NLTPKK52 pKa = 10.79GSTEE56 pKa = 4.02LPLSFVNTYY65 pKa = 10.91GFIKK69 pKa = 10.5PLLTYY74 pKa = 10.35SEE76 pKa = 5.41FYY78 pKa = 11.11SEE80 pKa = 4.24VHH82 pKa = 6.02HH83 pKa = 6.42QASTPCLTACMIPFGAGPYY102 pKa = 9.53IEE104 pKa = 4.61NPHH107 pKa = 6.93RR108 pKa = 11.84ILDD111 pKa = 3.79EE112 pKa = 4.25CDD114 pKa = 2.99KK115 pKa = 11.49VNIIVRR121 pKa = 11.84KK122 pKa = 8.64SASLKK127 pKa = 9.96EE128 pKa = 4.15EE129 pKa = 3.52IVFDD133 pKa = 4.05VRR135 pKa = 11.84RR136 pKa = 11.84LPPIFNKK143 pKa = 10.17HH144 pKa = 5.34QISGNRR150 pKa = 11.84LICVPSEE157 pKa = 4.41KK158 pKa = 10.2YY159 pKa = 10.28VKK161 pKa = 10.72SPGKK165 pKa = 9.29MVAGTDD171 pKa = 3.49YY172 pKa = 10.63AYY174 pKa = 10.84QIAFVSLTFCPEE186 pKa = 3.79SQKK189 pKa = 10.37FRR191 pKa = 11.84VARR194 pKa = 11.84PLQTIRR200 pKa = 11.84SPIMRR205 pKa = 11.84SVQLEE210 pKa = 3.89VMLRR214 pKa = 11.84IDD216 pKa = 4.14CAADD220 pKa = 3.58SPLKK224 pKa = 10.51RR225 pKa = 11.84LLIVNPGSTDD235 pKa = 3.22YY236 pKa = 11.12YY237 pKa = 9.65ASVWFHH243 pKa = 6.09ICNLYY248 pKa = 10.45RR249 pKa = 11.84GNKK252 pKa = 8.18PFKK255 pKa = 10.75SYY257 pKa = 11.47DD258 pKa = 3.28DD259 pKa = 4.02TYY261 pKa = 11.34FSQKK265 pKa = 8.99CRR267 pKa = 11.84AMQLEE272 pKa = 4.42CGIVDD277 pKa = 3.09MWGPTLVVKK286 pKa = 10.71AHH288 pKa = 5.64GKK290 pKa = 7.5IPKK293 pKa = 7.84MAKK296 pKa = 9.7PFFSAKK302 pKa = 9.05GWACHH307 pKa = 5.86AFSDD311 pKa = 5.38AAPTLAKK318 pKa = 10.59ALWSVGAKK326 pKa = 7.82ITQVNAILQPSEE338 pKa = 4.15LSQMVQVSDD347 pKa = 4.24VIWPKK352 pKa = 10.72VKK354 pKa = 10.44LDD356 pKa = 4.06NRR358 pKa = 11.84IQDD361 pKa = 3.41YY362 pKa = 10.98SAAKK366 pKa = 8.94WNPFKK371 pKa = 11.11KK372 pKa = 10.26PP373 pKa = 3.15
MM1 pKa = 8.01ALRR4 pKa = 11.84QATIPVDD11 pKa = 3.38VDD13 pKa = 3.34KK14 pKa = 11.42DD15 pKa = 3.94SEE17 pKa = 4.5KK18 pKa = 11.38NNLNPFPIIPVTRR31 pKa = 11.84DD32 pKa = 3.25DD33 pKa = 4.13GSSTGRR39 pKa = 11.84LVKK42 pKa = 9.77QLRR45 pKa = 11.84VKK47 pKa = 10.76NLTPKK52 pKa = 10.79GSTEE56 pKa = 4.02LPLSFVNTYY65 pKa = 10.91GFIKK69 pKa = 10.5PLLTYY74 pKa = 10.35SEE76 pKa = 5.41FYY78 pKa = 11.11SEE80 pKa = 4.24VHH82 pKa = 6.02HH83 pKa = 6.42QASTPCLTACMIPFGAGPYY102 pKa = 9.53IEE104 pKa = 4.61NPHH107 pKa = 6.93RR108 pKa = 11.84ILDD111 pKa = 3.79EE112 pKa = 4.25CDD114 pKa = 2.99KK115 pKa = 11.49VNIIVRR121 pKa = 11.84KK122 pKa = 8.64SASLKK127 pKa = 9.96EE128 pKa = 4.15EE129 pKa = 3.52IVFDD133 pKa = 4.05VRR135 pKa = 11.84RR136 pKa = 11.84LPPIFNKK143 pKa = 10.17HH144 pKa = 5.34QISGNRR150 pKa = 11.84LICVPSEE157 pKa = 4.41KK158 pKa = 10.2YY159 pKa = 10.28VKK161 pKa = 10.72SPGKK165 pKa = 9.29MVAGTDD171 pKa = 3.49YY172 pKa = 10.63AYY174 pKa = 10.84QIAFVSLTFCPEE186 pKa = 3.79SQKK189 pKa = 10.37FRR191 pKa = 11.84VARR194 pKa = 11.84PLQTIRR200 pKa = 11.84SPIMRR205 pKa = 11.84SVQLEE210 pKa = 3.89VMLRR214 pKa = 11.84IDD216 pKa = 4.14CAADD220 pKa = 3.58SPLKK224 pKa = 10.51RR225 pKa = 11.84LLIVNPGSTDD235 pKa = 3.22YY236 pKa = 11.12YY237 pKa = 9.65ASVWFHH243 pKa = 6.09ICNLYY248 pKa = 10.45RR249 pKa = 11.84GNKK252 pKa = 8.18PFKK255 pKa = 10.75SYY257 pKa = 11.47DD258 pKa = 3.28DD259 pKa = 4.02TYY261 pKa = 11.34FSQKK265 pKa = 8.99CRR267 pKa = 11.84AMQLEE272 pKa = 4.42CGIVDD277 pKa = 3.09MWGPTLVVKK286 pKa = 10.71AHH288 pKa = 5.64GKK290 pKa = 7.5IPKK293 pKa = 7.84MAKK296 pKa = 9.7PFFSAKK302 pKa = 9.05GWACHH307 pKa = 5.86AFSDD311 pKa = 5.38AAPTLAKK318 pKa = 10.59ALWSVGAKK326 pKa = 7.82ITQVNAILQPSEE338 pKa = 4.15LSQMVQVSDD347 pKa = 4.24VIWPKK352 pKa = 10.72VKK354 pKa = 10.44LDD356 pKa = 4.06NRR358 pKa = 11.84IQDD361 pKa = 3.41YY362 pKa = 10.98SAAKK366 pKa = 8.94WNPFKK371 pKa = 11.11KK372 pKa = 10.26PP373 pKa = 3.15
Molecular weight: 41.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4925 |
227 |
2269 |
703.6 |
78.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.97 ± 1.005 | 2.091 ± 0.251 |
5.36 ± 0.498 | 4.975 ± 0.572 |
3.655 ± 0.361 | 5.137 ± 0.601 |
1.949 ± 0.316 | 7.919 ± 0.734 |
5.015 ± 0.663 | 10.234 ± 1.191 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.843 ± 0.346 | 5.239 ± 0.47 |
5.178 ± 0.614 | 4.629 ± 0.673 |
4.954 ± 0.486 | 8.365 ± 0.634 |
6.477 ± 0.3 | 5.827 ± 0.613 |
1.056 ± 0.176 | 3.127 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
