
Rikenella microfusus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia;
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
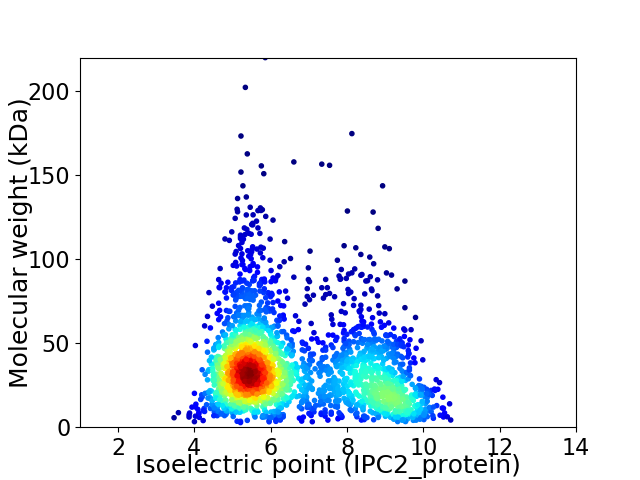
Virtual 2D-PAGE plot for 2358 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
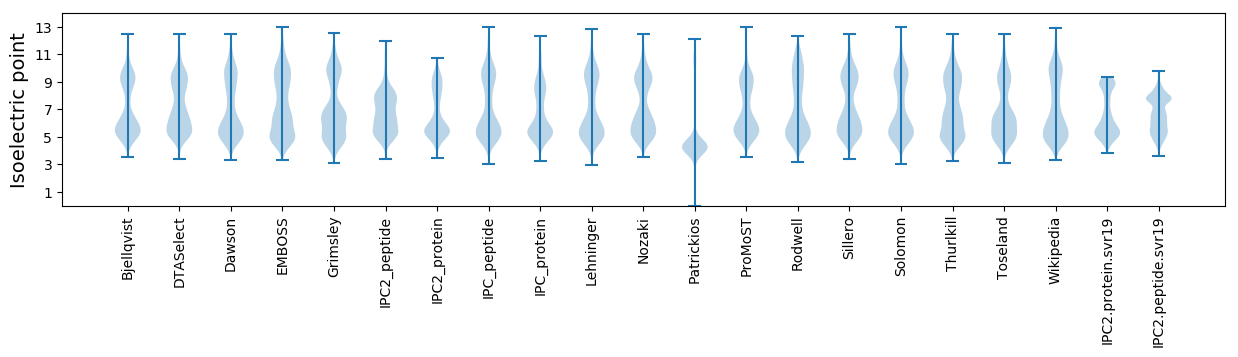
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A379MP92|A0A379MP92_9BACT Cd1 OS=Rikenella microfusus OX=28139 GN=NCTC11190_00753 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.49KK3 pKa = 10.28LFQFVCVAAIAAGFAACGGNAANTEE28 pKa = 4.33NADD31 pKa = 3.42STAVVEE37 pKa = 4.25MSEE40 pKa = 4.41TVVVDD45 pKa = 4.3TVAVDD50 pKa = 3.64TTACDD55 pKa = 3.44TTACDD60 pKa = 3.28SAAVAKK66 pKa = 10.58
MM1 pKa = 7.57KK2 pKa = 10.49KK3 pKa = 10.28LFQFVCVAAIAAGFAACGGNAANTEE28 pKa = 4.33NADD31 pKa = 3.42STAVVEE37 pKa = 4.25MSEE40 pKa = 4.41TVVVDD45 pKa = 4.3TVAVDD50 pKa = 3.64TTACDD55 pKa = 3.44TTACDD60 pKa = 3.28SAAVAKK66 pKa = 10.58
Molecular weight: 6.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A379MWB1|A0A379MWB1_9BACT Uncharacterized protein OS=Rikenella microfusus OX=28139 GN=NCTC11190_02383 PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 3.88KK3 pKa = 11.02HH4 pKa = 6.2PFLRR8 pKa = 11.84NFTNKK13 pKa = 9.33IPKK16 pKa = 9.97LLFLLAVLYY25 pKa = 10.65ALSQNKK31 pKa = 9.32GMLFGHH37 pKa = 6.26DD38 pKa = 4.03TEE40 pKa = 4.72PVRR43 pKa = 11.84ADD45 pKa = 3.29TPTPFDD51 pKa = 3.75TTCANAFFKK60 pKa = 10.89RR61 pKa = 11.84PVVLSTQDD69 pKa = 3.08SVLYY73 pKa = 9.37EE74 pKa = 3.98VRR76 pKa = 11.84FEE78 pKa = 5.11AGNDD82 pKa = 3.61DD83 pKa = 3.61PDD85 pKa = 5.57GYY87 pKa = 11.37VIAGKK92 pKa = 10.38RR93 pKa = 11.84KK94 pKa = 7.16TNSARR99 pKa = 11.84GFGGNVPVAVFLDD112 pKa = 3.71DD113 pKa = 3.73GMIIRR118 pKa = 11.84GVRR121 pKa = 11.84LLEE124 pKa = 4.28NNEE127 pKa = 3.85TPRR130 pKa = 11.84FLRR133 pKa = 11.84RR134 pKa = 11.84MDD136 pKa = 3.34EE137 pKa = 3.94EE138 pKa = 4.37RR139 pKa = 11.84FLEE142 pKa = 4.41RR143 pKa = 11.84WNGRR147 pKa = 11.84HH148 pKa = 6.49LYY150 pKa = 10.3DD151 pKa = 3.6PQKK154 pKa = 10.91RR155 pKa = 11.84IDD157 pKa = 3.69ASSGASYY164 pKa = 8.44TARR167 pKa = 11.84AVISNVNNTLAALLDD182 pKa = 4.38RR183 pKa = 11.84KK184 pKa = 9.66PLPPPRR190 pKa = 11.84RR191 pKa = 11.84FAADD195 pKa = 3.54YY196 pKa = 10.23LGEE199 pKa = 4.22SAVLLVTIMSLACFVSPQSTRR220 pKa = 11.84RR221 pKa = 11.84IRR223 pKa = 11.84IPLLLLSIAVLGIWQGAFLSLALLYY248 pKa = 10.35RR249 pKa = 11.84WLLFGASPLVRR260 pKa = 11.84FGIFAAVVLSLLLPFLTNRR279 pKa = 11.84RR280 pKa = 11.84FYY282 pKa = 10.75CAYY285 pKa = 10.0LCPFGAAQEE294 pKa = 4.12LAGRR298 pKa = 11.84LGMRR302 pKa = 11.84KK303 pKa = 7.86WAIPEE308 pKa = 3.89RR309 pKa = 11.84FLRR312 pKa = 11.84PARR315 pKa = 11.84WARR318 pKa = 11.84RR319 pKa = 11.84GILAAAVVLLLILPHH334 pKa = 6.7FEE336 pKa = 4.21LADD339 pKa = 3.56IEE341 pKa = 4.64PFTAFLIRR349 pKa = 11.84SASAASLTLAGLSLAASFFFQRR371 pKa = 11.84PWCRR375 pKa = 11.84LLCPTGEE382 pKa = 4.19LLSILRR388 pKa = 11.84RR389 pKa = 11.84PVRR392 pKa = 11.84YY393 pKa = 8.43PKK395 pKa = 10.24RR396 pKa = 11.84VLRR399 pKa = 11.84RR400 pKa = 11.84NPPGCRR406 pKa = 3.24
MM1 pKa = 8.12DD2 pKa = 3.88KK3 pKa = 11.02HH4 pKa = 6.2PFLRR8 pKa = 11.84NFTNKK13 pKa = 9.33IPKK16 pKa = 9.97LLFLLAVLYY25 pKa = 10.65ALSQNKK31 pKa = 9.32GMLFGHH37 pKa = 6.26DD38 pKa = 4.03TEE40 pKa = 4.72PVRR43 pKa = 11.84ADD45 pKa = 3.29TPTPFDD51 pKa = 3.75TTCANAFFKK60 pKa = 10.89RR61 pKa = 11.84PVVLSTQDD69 pKa = 3.08SVLYY73 pKa = 9.37EE74 pKa = 3.98VRR76 pKa = 11.84FEE78 pKa = 5.11AGNDD82 pKa = 3.61DD83 pKa = 3.61PDD85 pKa = 5.57GYY87 pKa = 11.37VIAGKK92 pKa = 10.38RR93 pKa = 11.84KK94 pKa = 7.16TNSARR99 pKa = 11.84GFGGNVPVAVFLDD112 pKa = 3.71DD113 pKa = 3.73GMIIRR118 pKa = 11.84GVRR121 pKa = 11.84LLEE124 pKa = 4.28NNEE127 pKa = 3.85TPRR130 pKa = 11.84FLRR133 pKa = 11.84RR134 pKa = 11.84MDD136 pKa = 3.34EE137 pKa = 3.94EE138 pKa = 4.37RR139 pKa = 11.84FLEE142 pKa = 4.41RR143 pKa = 11.84WNGRR147 pKa = 11.84HH148 pKa = 6.49LYY150 pKa = 10.3DD151 pKa = 3.6PQKK154 pKa = 10.91RR155 pKa = 11.84IDD157 pKa = 3.69ASSGASYY164 pKa = 8.44TARR167 pKa = 11.84AVISNVNNTLAALLDD182 pKa = 4.38RR183 pKa = 11.84KK184 pKa = 9.66PLPPPRR190 pKa = 11.84RR191 pKa = 11.84FAADD195 pKa = 3.54YY196 pKa = 10.23LGEE199 pKa = 4.22SAVLLVTIMSLACFVSPQSTRR220 pKa = 11.84RR221 pKa = 11.84IRR223 pKa = 11.84IPLLLLSIAVLGIWQGAFLSLALLYY248 pKa = 10.35RR249 pKa = 11.84WLLFGASPLVRR260 pKa = 11.84FGIFAAVVLSLLLPFLTNRR279 pKa = 11.84RR280 pKa = 11.84FYY282 pKa = 10.75CAYY285 pKa = 10.0LCPFGAAQEE294 pKa = 4.12LAGRR298 pKa = 11.84LGMRR302 pKa = 11.84KK303 pKa = 7.86WAIPEE308 pKa = 3.89RR309 pKa = 11.84FLRR312 pKa = 11.84PARR315 pKa = 11.84WARR318 pKa = 11.84RR319 pKa = 11.84GILAAAVVLLLILPHH334 pKa = 6.7FEE336 pKa = 4.21LADD339 pKa = 3.56IEE341 pKa = 4.64PFTAFLIRR349 pKa = 11.84SASAASLTLAGLSLAASFFFQRR371 pKa = 11.84PWCRR375 pKa = 11.84LLCPTGEE382 pKa = 4.19LLSILRR388 pKa = 11.84RR389 pKa = 11.84PVRR392 pKa = 11.84YY393 pKa = 8.43PKK395 pKa = 10.24RR396 pKa = 11.84VLRR399 pKa = 11.84RR400 pKa = 11.84NPPGCRR406 pKa = 3.24
Molecular weight: 45.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
804626 |
29 |
1938 |
341.2 |
37.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.404 ± 0.059 | 1.131 ± 0.022 |
5.607 ± 0.038 | 6.355 ± 0.046 |
4.201 ± 0.032 | 7.915 ± 0.04 |
1.76 ± 0.023 | 5.86 ± 0.041 |
4.775 ± 0.056 | 9.112 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.531 ± 0.019 | 3.72 ± 0.036 |
4.487 ± 0.034 | 3.079 ± 0.025 |
6.655 ± 0.05 | 5.536 ± 0.036 |
5.712 ± 0.036 | 7.087 ± 0.037 |
1.267 ± 0.023 | 3.805 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
