
Isoptericola variabilis (strain 225)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Isoptericola; Isoptericola variabilis
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
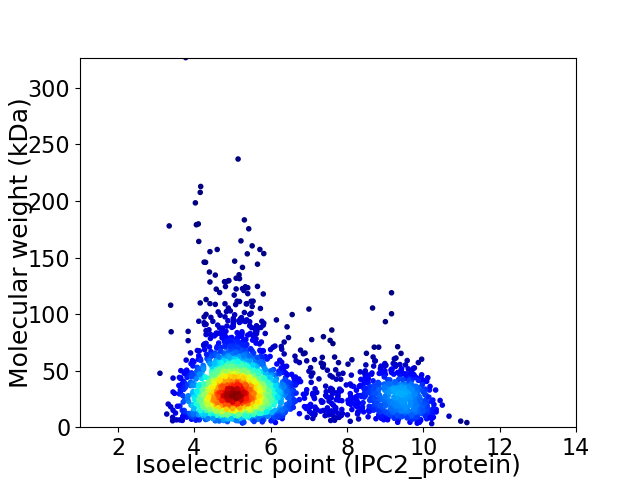
Virtual 2D-PAGE plot for 2879 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6FPW0|F6FPW0_ISOV2 Monosaccharide-transporting ATPase OS=Isoptericola variabilis (strain 225) OX=743718 GN=Isova_0965 PE=4 SV=1
MM1 pKa = 7.0TPRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84LAGIGAFALASALVLSACGGGGGDD30 pKa = 3.67NTDD33 pKa = 3.28TGEE36 pKa = 4.45GGDD39 pKa = 3.67AGATEE44 pKa = 5.17GGATTGIVTVNGTEE58 pKa = 4.04PQNPLTPGMTNEE70 pKa = 4.19VGGGLAVKK78 pKa = 10.49NIFAGLVYY86 pKa = 10.5YY87 pKa = 10.43DD88 pKa = 4.37AEE90 pKa = 4.65GATHH94 pKa = 6.69YY95 pKa = 11.13EE96 pKa = 3.93MAEE99 pKa = 4.21SIEE102 pKa = 4.98SEE104 pKa = 4.9DD105 pKa = 3.67NQNWTITIQDD115 pKa = 3.05GWTFSDD121 pKa = 3.93GTPVTAQSYY130 pKa = 8.82VDD132 pKa = 3.19AWKK135 pKa = 8.91YY136 pKa = 8.08TALLSNGQNQQYY148 pKa = 9.07FFHH151 pKa = 7.6DD152 pKa = 3.9FAGWSEE158 pKa = 4.4TEE160 pKa = 3.98DD161 pKa = 3.51SEE163 pKa = 4.94IAVEE167 pKa = 4.42VVDD170 pKa = 4.46DD171 pKa = 3.71HH172 pKa = 6.4TFTVEE177 pKa = 3.79LAFPNAEE184 pKa = 4.1FEE186 pKa = 4.3RR187 pKa = 11.84TLGYY191 pKa = 8.34TAFMPMPEE199 pKa = 4.46VFFEE203 pKa = 4.4TPDD206 pKa = 3.27IYY208 pKa = 11.41SDD210 pKa = 3.76APVGNGPYY218 pKa = 9.95VVEE221 pKa = 4.27SWEE224 pKa = 4.15HH225 pKa = 5.68EE226 pKa = 4.15NVISLRR232 pKa = 11.84PNPDD236 pKa = 2.82YY237 pKa = 10.84TGPRR241 pKa = 11.84TPQNGGIDD249 pKa = 3.22IVIYY253 pKa = 10.17TDD255 pKa = 3.53EE256 pKa = 4.02NTAYY260 pKa = 11.08NDD262 pKa = 4.03LLSDD266 pKa = 3.78NLDD269 pKa = 3.77IITNVPASAFQTFEE283 pKa = 3.9SDD285 pKa = 3.3LGDD288 pKa = 3.35RR289 pKa = 11.84AVNQPAAVIQVIQVPAWLPEE309 pKa = 4.01FEE311 pKa = 5.09GEE313 pKa = 3.96AGNLRR318 pKa = 11.84RR319 pKa = 11.84QAISKK324 pKa = 10.61AIDD327 pKa = 3.2RR328 pKa = 11.84EE329 pKa = 4.48QVTEE333 pKa = 4.15TVFAGTRR340 pKa = 11.84TPATDD345 pKa = 3.5FTSPVIDD352 pKa = 5.19GYY354 pKa = 11.43SEE356 pKa = 5.76DD357 pKa = 3.72IPGSEE362 pKa = 4.11VLQYY366 pKa = 11.01DD367 pKa = 3.74PEE369 pKa = 4.57GAKK372 pKa = 9.7QLWDD376 pKa = 3.58EE377 pKa = 4.61AEE379 pKa = 4.31GMDD382 pKa = 4.84PLGDD386 pKa = 3.64YY387 pKa = 8.89TLQLASNADD396 pKa = 3.64SDD398 pKa = 3.84HH399 pKa = 5.24QTWVEE404 pKa = 3.87AVCNNIRR411 pKa = 11.84TVLEE415 pKa = 4.32IEE417 pKa = 4.57CEE419 pKa = 4.25FYY421 pKa = 10.41PYY423 pKa = 9.02PTFAEE428 pKa = 4.22FLDD431 pKa = 3.87ARR433 pKa = 11.84DD434 pKa = 3.69AGEE437 pKa = 4.02VPGLFRR443 pKa = 11.84GGWQADD449 pKa = 3.56WPAMSNFLGPVYY461 pKa = 10.0GTGAGSNDD469 pKa = 3.01MQYY472 pKa = 11.53SNEE475 pKa = 3.98EE476 pKa = 3.49FDD478 pKa = 5.13AKK480 pKa = 10.02LTEE483 pKa = 4.45AGAAEE488 pKa = 4.33SPEE491 pKa = 4.26EE492 pKa = 3.73ATEE495 pKa = 4.37LYY497 pKa = 10.66KK498 pKa = 10.52EE499 pKa = 4.04AQEE502 pKa = 4.07ILFRR506 pKa = 11.84DD507 pKa = 3.95LPGIPLWYY515 pKa = 10.49NNATAGYY522 pKa = 9.13AATVDD527 pKa = 3.48NVQFGWDD534 pKa = 3.64SDD536 pKa = 4.75PILHH540 pKa = 6.56EE541 pKa = 4.3VTKK544 pKa = 11.05SEE546 pKa = 4.04
MM1 pKa = 7.0TPRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84LAGIGAFALASALVLSACGGGGGDD30 pKa = 3.67NTDD33 pKa = 3.28TGEE36 pKa = 4.45GGDD39 pKa = 3.67AGATEE44 pKa = 5.17GGATTGIVTVNGTEE58 pKa = 4.04PQNPLTPGMTNEE70 pKa = 4.19VGGGLAVKK78 pKa = 10.49NIFAGLVYY86 pKa = 10.5YY87 pKa = 10.43DD88 pKa = 4.37AEE90 pKa = 4.65GATHH94 pKa = 6.69YY95 pKa = 11.13EE96 pKa = 3.93MAEE99 pKa = 4.21SIEE102 pKa = 4.98SEE104 pKa = 4.9DD105 pKa = 3.67NQNWTITIQDD115 pKa = 3.05GWTFSDD121 pKa = 3.93GTPVTAQSYY130 pKa = 8.82VDD132 pKa = 3.19AWKK135 pKa = 8.91YY136 pKa = 8.08TALLSNGQNQQYY148 pKa = 9.07FFHH151 pKa = 7.6DD152 pKa = 3.9FAGWSEE158 pKa = 4.4TEE160 pKa = 3.98DD161 pKa = 3.51SEE163 pKa = 4.94IAVEE167 pKa = 4.42VVDD170 pKa = 4.46DD171 pKa = 3.71HH172 pKa = 6.4TFTVEE177 pKa = 3.79LAFPNAEE184 pKa = 4.1FEE186 pKa = 4.3RR187 pKa = 11.84TLGYY191 pKa = 8.34TAFMPMPEE199 pKa = 4.46VFFEE203 pKa = 4.4TPDD206 pKa = 3.27IYY208 pKa = 11.41SDD210 pKa = 3.76APVGNGPYY218 pKa = 9.95VVEE221 pKa = 4.27SWEE224 pKa = 4.15HH225 pKa = 5.68EE226 pKa = 4.15NVISLRR232 pKa = 11.84PNPDD236 pKa = 2.82YY237 pKa = 10.84TGPRR241 pKa = 11.84TPQNGGIDD249 pKa = 3.22IVIYY253 pKa = 10.17TDD255 pKa = 3.53EE256 pKa = 4.02NTAYY260 pKa = 11.08NDD262 pKa = 4.03LLSDD266 pKa = 3.78NLDD269 pKa = 3.77IITNVPASAFQTFEE283 pKa = 3.9SDD285 pKa = 3.3LGDD288 pKa = 3.35RR289 pKa = 11.84AVNQPAAVIQVIQVPAWLPEE309 pKa = 4.01FEE311 pKa = 5.09GEE313 pKa = 3.96AGNLRR318 pKa = 11.84RR319 pKa = 11.84QAISKK324 pKa = 10.61AIDD327 pKa = 3.2RR328 pKa = 11.84EE329 pKa = 4.48QVTEE333 pKa = 4.15TVFAGTRR340 pKa = 11.84TPATDD345 pKa = 3.5FTSPVIDD352 pKa = 5.19GYY354 pKa = 11.43SEE356 pKa = 5.76DD357 pKa = 3.72IPGSEE362 pKa = 4.11VLQYY366 pKa = 11.01DD367 pKa = 3.74PEE369 pKa = 4.57GAKK372 pKa = 9.7QLWDD376 pKa = 3.58EE377 pKa = 4.61AEE379 pKa = 4.31GMDD382 pKa = 4.84PLGDD386 pKa = 3.64YY387 pKa = 8.89TLQLASNADD396 pKa = 3.64SDD398 pKa = 3.84HH399 pKa = 5.24QTWVEE404 pKa = 3.87AVCNNIRR411 pKa = 11.84TVLEE415 pKa = 4.32IEE417 pKa = 4.57CEE419 pKa = 4.25FYY421 pKa = 10.41PYY423 pKa = 9.02PTFAEE428 pKa = 4.22FLDD431 pKa = 3.87ARR433 pKa = 11.84DD434 pKa = 3.69AGEE437 pKa = 4.02VPGLFRR443 pKa = 11.84GGWQADD449 pKa = 3.56WPAMSNFLGPVYY461 pKa = 10.0GTGAGSNDD469 pKa = 3.01MQYY472 pKa = 11.53SNEE475 pKa = 3.98EE476 pKa = 3.49FDD478 pKa = 5.13AKK480 pKa = 10.02LTEE483 pKa = 4.45AGAAEE488 pKa = 4.33SPEE491 pKa = 4.26EE492 pKa = 3.73ATEE495 pKa = 4.37LYY497 pKa = 10.66KK498 pKa = 10.52EE499 pKa = 4.04AQEE502 pKa = 4.07ILFRR506 pKa = 11.84DD507 pKa = 3.95LPGIPLWYY515 pKa = 10.49NNATAGYY522 pKa = 9.13AATVDD527 pKa = 3.48NVQFGWDD534 pKa = 3.64SDD536 pKa = 4.75PILHH540 pKa = 6.56EE541 pKa = 4.3VTKK544 pKa = 11.05SEE546 pKa = 4.04
Molecular weight: 59.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6FV08|F6FV08_ISOV2 Fmu (Sun) domain protein OS=Isoptericola variabilis (strain 225) OX=743718 GN=Isova_1595 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
987482 |
30 |
3247 |
343.0 |
36.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.278 ± 0.077 | 0.541 ± 0.01 |
6.508 ± 0.045 | 5.985 ± 0.045 |
2.593 ± 0.028 | 9.355 ± 0.049 |
2.151 ± 0.023 | 2.798 ± 0.027 |
1.513 ± 0.026 | 10.059 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.591 ± 0.015 | 1.522 ± 0.022 |
6.057 ± 0.035 | 2.495 ± 0.023 |
7.983 ± 0.049 | 4.702 ± 0.029 |
6.19 ± 0.039 | 10.253 ± 0.053 |
1.551 ± 0.018 | 1.873 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
