
Cohaesibacter marisflavi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Cohaesibacteraceae; Cohaesibacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
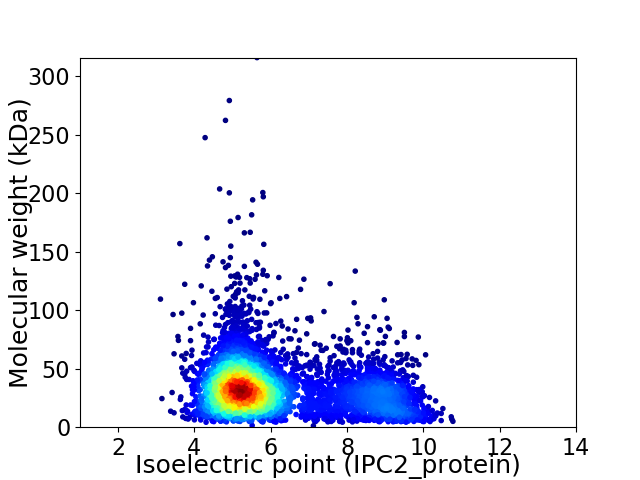
Virtual 2D-PAGE plot for 4794 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I5CN02|A0A1I5CN02_9RHIZ 2-dehydro-3-deoxygluconokinase OS=Cohaesibacter marisflavi OX=655353 GN=SAMN04488056_102284 PE=4 SV=1
MM1 pKa = 8.03SLMGATNAAITGLAAQSQALSNVSNNLANSDD32 pKa = 3.42TTAYY36 pKa = 9.47KK37 pKa = 10.47ASKK40 pKa = 9.36TSFSSLVAGSGNNKK54 pKa = 9.96SGGVHH59 pKa = 7.49ASNTTNNTAQGLVTDD74 pKa = 4.67SAISTHH80 pKa = 6.83LAIQGNGYY88 pKa = 9.63FVVSEE93 pKa = 4.33EE94 pKa = 4.58GDD96 pKa = 3.5TSSRR100 pKa = 11.84YY101 pKa = 5.33YY102 pKa = 9.63TRR104 pKa = 11.84NGEE107 pKa = 4.07FAVDD111 pKa = 3.22NDD113 pKa = 4.33GYY115 pKa = 11.53LMNGDD120 pKa = 4.15YY121 pKa = 11.13YY122 pKa = 11.54LLGWATDD129 pKa = 3.25ADD131 pKa = 4.31GNVAGGASSVNLDD144 pKa = 4.27KK145 pKa = 10.82IDD147 pKa = 4.05LSSIQSSAQATSEE160 pKa = 3.95ASIQATLPGDD170 pKa = 3.42AVLNSTFEE178 pKa = 4.31SSFEE182 pKa = 4.34VYY184 pKa = 10.48DD185 pKa = 3.88SLGTASTITATYY197 pKa = 9.7EE198 pKa = 3.65KK199 pKa = 9.43TAQNTWTITYY209 pKa = 9.32SDD211 pKa = 4.5PVSADD216 pKa = 2.95TSTTTGSVTSSPITVQFNNDD236 pKa = 2.45GSLASVSGTDD246 pKa = 6.22LDD248 pKa = 4.1ITWNTGAAASSIKK261 pKa = 10.45LDD263 pKa = 3.63LGTVGGSDD271 pKa = 3.87GLTQYY276 pKa = 11.18SSNDD280 pKa = 3.3VEE282 pKa = 4.58NADD285 pKa = 3.95IDD287 pKa = 4.17VKK289 pKa = 11.16SIIVDD294 pKa = 3.35GRR296 pKa = 11.84AYY298 pKa = 9.87GTVDD302 pKa = 4.43DD303 pKa = 5.79IEE305 pKa = 5.79IEE307 pKa = 4.14TDD309 pKa = 3.33GSVVATFSNGEE320 pKa = 3.66TRR322 pKa = 11.84NIYY325 pKa = 9.7KK326 pKa = 10.2IPVATFINSDD336 pKa = 3.55GLTEE340 pKa = 4.78DD341 pKa = 3.34SDD343 pKa = 4.46GIYY346 pKa = 10.53AISTYY351 pKa = 10.76SGNATLHH358 pKa = 5.4TAGTGSAGSLKK369 pKa = 10.58SSSLEE374 pKa = 3.84SSTVDD379 pKa = 2.9TSTEE383 pKa = 3.95FASMLSAQQAYY394 pKa = 10.4SSASQIISTAGDD406 pKa = 3.45MFDD409 pKa = 5.09SLLQAVRR416 pKa = 3.68
MM1 pKa = 8.03SLMGATNAAITGLAAQSQALSNVSNNLANSDD32 pKa = 3.42TTAYY36 pKa = 9.47KK37 pKa = 10.47ASKK40 pKa = 9.36TSFSSLVAGSGNNKK54 pKa = 9.96SGGVHH59 pKa = 7.49ASNTTNNTAQGLVTDD74 pKa = 4.67SAISTHH80 pKa = 6.83LAIQGNGYY88 pKa = 9.63FVVSEE93 pKa = 4.33EE94 pKa = 4.58GDD96 pKa = 3.5TSSRR100 pKa = 11.84YY101 pKa = 5.33YY102 pKa = 9.63TRR104 pKa = 11.84NGEE107 pKa = 4.07FAVDD111 pKa = 3.22NDD113 pKa = 4.33GYY115 pKa = 11.53LMNGDD120 pKa = 4.15YY121 pKa = 11.13YY122 pKa = 11.54LLGWATDD129 pKa = 3.25ADD131 pKa = 4.31GNVAGGASSVNLDD144 pKa = 4.27KK145 pKa = 10.82IDD147 pKa = 4.05LSSIQSSAQATSEE160 pKa = 3.95ASIQATLPGDD170 pKa = 3.42AVLNSTFEE178 pKa = 4.31SSFEE182 pKa = 4.34VYY184 pKa = 10.48DD185 pKa = 3.88SLGTASTITATYY197 pKa = 9.7EE198 pKa = 3.65KK199 pKa = 9.43TAQNTWTITYY209 pKa = 9.32SDD211 pKa = 4.5PVSADD216 pKa = 2.95TSTTTGSVTSSPITVQFNNDD236 pKa = 2.45GSLASVSGTDD246 pKa = 6.22LDD248 pKa = 4.1ITWNTGAAASSIKK261 pKa = 10.45LDD263 pKa = 3.63LGTVGGSDD271 pKa = 3.87GLTQYY276 pKa = 11.18SSNDD280 pKa = 3.3VEE282 pKa = 4.58NADD285 pKa = 3.95IDD287 pKa = 4.17VKK289 pKa = 11.16SIIVDD294 pKa = 3.35GRR296 pKa = 11.84AYY298 pKa = 9.87GTVDD302 pKa = 4.43DD303 pKa = 5.79IEE305 pKa = 5.79IEE307 pKa = 4.14TDD309 pKa = 3.33GSVVATFSNGEE320 pKa = 3.66TRR322 pKa = 11.84NIYY325 pKa = 9.7KK326 pKa = 10.2IPVATFINSDD336 pKa = 3.55GLTEE340 pKa = 4.78DD341 pKa = 3.34SDD343 pKa = 4.46GIYY346 pKa = 10.53AISTYY351 pKa = 10.76SGNATLHH358 pKa = 5.4TAGTGSAGSLKK369 pKa = 10.58SSSLEE374 pKa = 3.84SSTVDD379 pKa = 2.9TSTEE383 pKa = 3.95FASMLSAQQAYY394 pKa = 10.4SSASQIISTAGDD406 pKa = 3.45MFDD409 pKa = 5.09SLLQAVRR416 pKa = 3.68
Molecular weight: 42.69 kDa
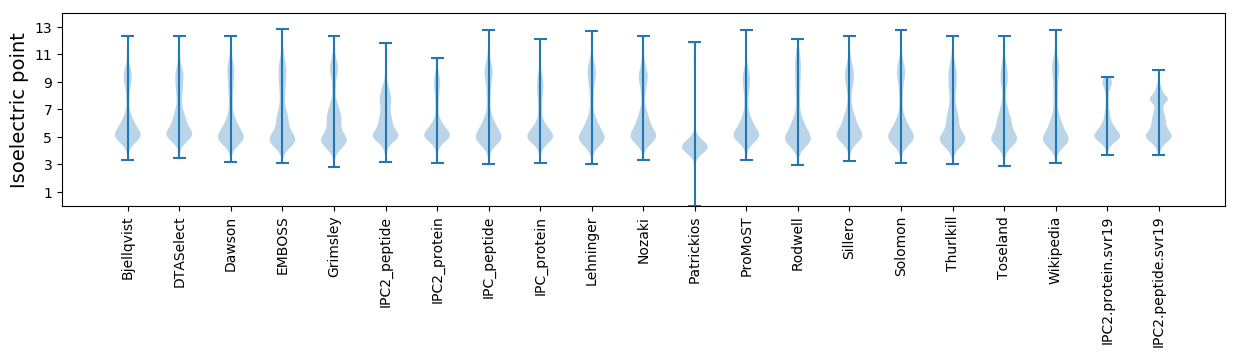
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I5F2R4|A0A1I5F2R4_9RHIZ Transcriptional regulator AraC family OS=Cohaesibacter marisflavi OX=655353 GN=SAMN04488056_103462 PE=4 SV=1
MM1 pKa = 8.21DD2 pKa = 4.9MFPEE6 pKa = 3.92KK7 pKa = 10.38AARR10 pKa = 11.84FKK12 pKa = 10.99GIRR15 pKa = 11.84AGASTSFFTAALGSIVSRR33 pKa = 11.84TSARR37 pKa = 11.84GAVHH41 pKa = 6.52PRR43 pKa = 11.84LSGHH47 pKa = 5.7SLYY50 pKa = 10.57LRR52 pKa = 11.84HH53 pKa = 6.16PVKK56 pKa = 10.77GDD58 pKa = 3.44FAQWVRR64 pKa = 11.84LRR66 pKa = 11.84TASAGFLKK74 pKa = 10.08PWEE77 pKa = 4.3PLWPRR82 pKa = 11.84DD83 pKa = 3.68DD84 pKa = 3.5LTMRR88 pKa = 11.84GYY90 pKa = 10.65RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84LDD95 pKa = 3.05QYY97 pKa = 11.09RR98 pKa = 11.84RR99 pKa = 11.84DD100 pKa = 3.84RR101 pKa = 11.84KK102 pKa = 8.44TGRR105 pKa = 11.84ALPYY109 pKa = 10.27LIFDD113 pKa = 3.84ARR115 pKa = 11.84DD116 pKa = 3.12NRR118 pKa = 11.84LLGGVNVSNIRR129 pKa = 11.84RR130 pKa = 11.84GICQTATIGYY140 pKa = 8.96WMGEE144 pKa = 3.78NHH146 pKa = 7.23AGQGNMSRR154 pKa = 11.84ALALLLPYY162 pKa = 10.85LFDD165 pKa = 3.85CQGLHH170 pKa = 6.98RR171 pKa = 11.84IEE173 pKa = 4.6AACLPTNSASISVLKK188 pKa = 10.8KK189 pKa = 10.66SGFQLEE195 pKa = 4.54GKK197 pKa = 9.61ARR199 pKa = 11.84SYY201 pKa = 11.56LCINGQWEE209 pKa = 4.1DD210 pKa = 3.6HH211 pKa = 6.55LLFSALRR218 pKa = 11.84SDD220 pKa = 4.37VIMSVGFF227 pKa = 3.59
MM1 pKa = 8.21DD2 pKa = 4.9MFPEE6 pKa = 3.92KK7 pKa = 10.38AARR10 pKa = 11.84FKK12 pKa = 10.99GIRR15 pKa = 11.84AGASTSFFTAALGSIVSRR33 pKa = 11.84TSARR37 pKa = 11.84GAVHH41 pKa = 6.52PRR43 pKa = 11.84LSGHH47 pKa = 5.7SLYY50 pKa = 10.57LRR52 pKa = 11.84HH53 pKa = 6.16PVKK56 pKa = 10.77GDD58 pKa = 3.44FAQWVRR64 pKa = 11.84LRR66 pKa = 11.84TASAGFLKK74 pKa = 10.08PWEE77 pKa = 4.3PLWPRR82 pKa = 11.84DD83 pKa = 3.68DD84 pKa = 3.5LTMRR88 pKa = 11.84GYY90 pKa = 10.65RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84LDD95 pKa = 3.05QYY97 pKa = 11.09RR98 pKa = 11.84RR99 pKa = 11.84DD100 pKa = 3.84RR101 pKa = 11.84KK102 pKa = 8.44TGRR105 pKa = 11.84ALPYY109 pKa = 10.27LIFDD113 pKa = 3.84ARR115 pKa = 11.84DD116 pKa = 3.12NRR118 pKa = 11.84LLGGVNVSNIRR129 pKa = 11.84RR130 pKa = 11.84GICQTATIGYY140 pKa = 8.96WMGEE144 pKa = 3.78NHH146 pKa = 7.23AGQGNMSRR154 pKa = 11.84ALALLLPYY162 pKa = 10.85LFDD165 pKa = 3.85CQGLHH170 pKa = 6.98RR171 pKa = 11.84IEE173 pKa = 4.6AACLPTNSASISVLKK188 pKa = 10.8KK189 pKa = 10.66SGFQLEE195 pKa = 4.54GKK197 pKa = 9.61ARR199 pKa = 11.84SYY201 pKa = 11.56LCINGQWEE209 pKa = 4.1DD210 pKa = 3.6HH211 pKa = 6.55LLFSALRR218 pKa = 11.84SDD220 pKa = 4.37VIMSVGFF227 pKa = 3.59
Molecular weight: 25.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1536085 |
10 |
2835 |
320.4 |
35.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.547 ± 0.038 | 0.942 ± 0.012 |
5.92 ± 0.033 | 6.174 ± 0.038 |
3.999 ± 0.022 | 7.912 ± 0.039 |
2.053 ± 0.016 | 6.024 ± 0.027 |
4.529 ± 0.027 | 10.1 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.86 ± 0.017 | 3.198 ± 0.02 |
4.556 ± 0.028 | 3.476 ± 0.02 |
5.561 ± 0.03 | 6.346 ± 0.031 |
5.215 ± 0.021 | 6.906 ± 0.027 |
1.218 ± 0.016 | 2.465 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
