
Hubei sobemo-like virus 44
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
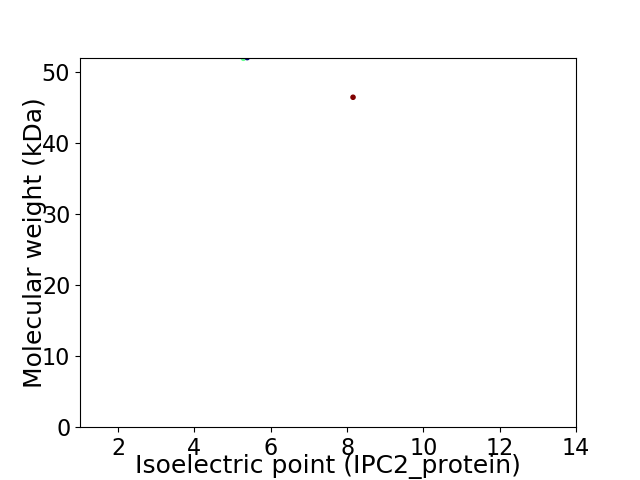
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
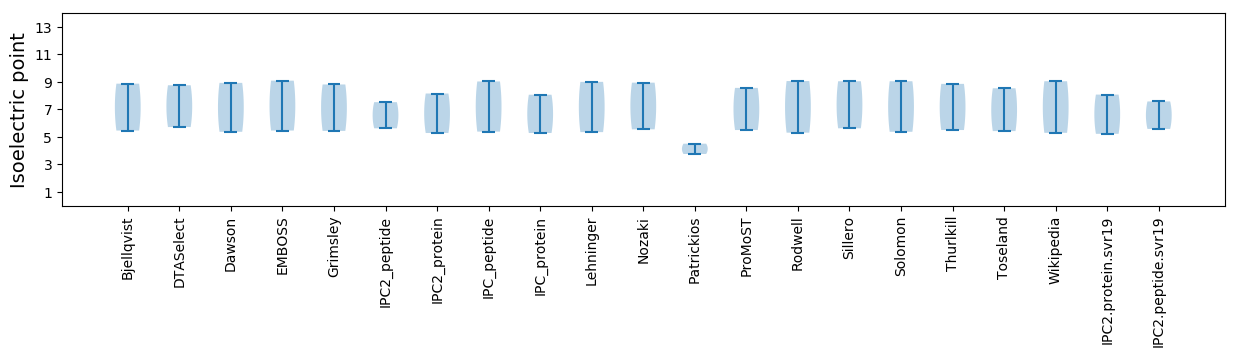
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KET7|A0A1L3KET7_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 44 OX=1923232 PE=4 SV=1
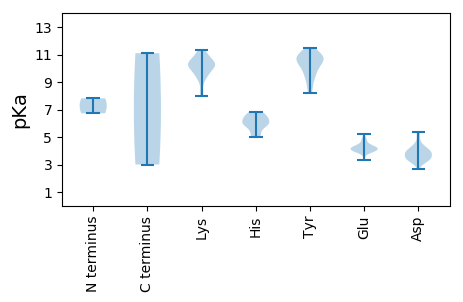
MM1 pKa = 6.73EE2 pKa = 4.54TLRR5 pKa = 11.84RR6 pKa = 11.84AGQALEE12 pKa = 4.24EE13 pKa = 4.51TTSEE17 pKa = 4.21TVNGIKK23 pKa = 10.34SLLGAVSDD31 pKa = 4.66CLEE34 pKa = 4.14EE35 pKa = 4.3FQWPHH40 pKa = 5.85NADD43 pKa = 3.8DD44 pKa = 3.91LTSFRR49 pKa = 11.84IHH51 pKa = 6.45ADD53 pKa = 2.68KK54 pKa = 11.0YY55 pKa = 10.92FRR57 pKa = 11.84LATNACIPDD66 pKa = 4.1LEE68 pKa = 4.27LQEE71 pKa = 4.97RR72 pKa = 11.84VLNKK76 pKa = 9.36TEE78 pKa = 3.35EE79 pKa = 4.2WYY81 pKa = 10.83APVRR85 pKa = 11.84WKK87 pKa = 11.19LPDD90 pKa = 3.36NYY92 pKa = 10.65LSKK95 pKa = 10.98EE96 pKa = 4.02VFKK99 pKa = 10.68TVLRR103 pKa = 11.84RR104 pKa = 11.84LDD106 pKa = 3.94FSSSPGYY113 pKa = 9.37PYY115 pKa = 10.65CKK117 pKa = 9.28RR118 pKa = 11.84KK119 pKa = 8.01QTIGEE124 pKa = 3.98WLGFDD129 pKa = 4.42GFQFDD134 pKa = 4.22PVQVDD139 pKa = 4.51LLWMDD144 pKa = 3.35VQRR147 pKa = 11.84AILGEE152 pKa = 4.18SEE154 pKa = 4.83LIMRR158 pKa = 11.84VFIKK162 pKa = 10.42QEE164 pKa = 3.47PHH166 pKa = 5.9KK167 pKa = 10.44AAKK170 pKa = 9.93AVEE173 pKa = 4.26GRR175 pKa = 11.84WRR177 pKa = 11.84LIMAFPLAHH186 pKa = 5.15QVLWHH191 pKa = 5.86MLFDD195 pKa = 3.9YY196 pKa = 10.95MNDD199 pKa = 3.52LEE201 pKa = 4.32ITKK204 pKa = 10.21AAEE207 pKa = 3.87IPSQQGIVLYY217 pKa = 10.59GGAWKK222 pKa = 10.25SHH224 pKa = 5.29LARR227 pKa = 11.84WKK229 pKa = 9.3QQGYY233 pKa = 8.17DD234 pKa = 3.13TGLDD238 pKa = 3.15KK239 pKa = 11.3SAWDD243 pKa = 3.15WTYY246 pKa = 10.81TYY248 pKa = 11.07WLMQMDD254 pKa = 4.45LQLRR258 pKa = 11.84YY259 pKa = 8.79RR260 pKa = 11.84TGRR263 pKa = 11.84GFKK266 pKa = 9.85MMEE269 pKa = 3.92WFQLATRR276 pKa = 11.84EE277 pKa = 4.03YY278 pKa = 9.29EE279 pKa = 3.94LAFGVGAKK287 pKa = 10.4FITTDD292 pKa = 3.53GYY294 pKa = 11.06EE295 pKa = 4.15LEE297 pKa = 4.15QLVPGLMKK305 pKa = 10.49SGSVNTISSNSHH317 pKa = 4.99AQAMLHH323 pKa = 6.24ALVCIEE329 pKa = 4.74GGVDD333 pKa = 3.53PDD335 pKa = 5.21PYY337 pKa = 10.23PACCGDD343 pKa = 3.68DD344 pKa = 3.67TLQKK348 pKa = 9.85MSQALIPLYY357 pKa = 9.91EE358 pKa = 4.38KK359 pKa = 10.76YY360 pKa = 10.92GVVVKK365 pKa = 10.55SASEE369 pKa = 3.76GMEE372 pKa = 4.49FIGHH376 pKa = 6.57EE377 pKa = 4.34FLDD380 pKa = 4.22SGPQPLYY387 pKa = 9.82MRR389 pKa = 11.84KK390 pKa = 8.66HH391 pKa = 6.0LNRR394 pKa = 11.84FLHH397 pKa = 6.28MKK399 pKa = 10.45DD400 pKa = 3.94EE401 pKa = 4.18YY402 pKa = 11.38LGDD405 pKa = 3.89YY406 pKa = 10.09LDD408 pKa = 4.2GMSRR412 pKa = 11.84LYY414 pKa = 11.16AKK416 pKa = 9.56TPYY419 pKa = 10.29FEE421 pKa = 4.04FWSWLGDD428 pKa = 3.72VYY430 pKa = 11.26DD431 pKa = 4.14VPVPSKK437 pKa = 10.72QYY439 pKa = 9.19VNNWYY444 pKa = 9.77DD445 pKa = 2.94HH446 pKa = 6.8SYY448 pKa = 11.1
MM1 pKa = 6.73EE2 pKa = 4.54TLRR5 pKa = 11.84RR6 pKa = 11.84AGQALEE12 pKa = 4.24EE13 pKa = 4.51TTSEE17 pKa = 4.21TVNGIKK23 pKa = 10.34SLLGAVSDD31 pKa = 4.66CLEE34 pKa = 4.14EE35 pKa = 4.3FQWPHH40 pKa = 5.85NADD43 pKa = 3.8DD44 pKa = 3.91LTSFRR49 pKa = 11.84IHH51 pKa = 6.45ADD53 pKa = 2.68KK54 pKa = 11.0YY55 pKa = 10.92FRR57 pKa = 11.84LATNACIPDD66 pKa = 4.1LEE68 pKa = 4.27LQEE71 pKa = 4.97RR72 pKa = 11.84VLNKK76 pKa = 9.36TEE78 pKa = 3.35EE79 pKa = 4.2WYY81 pKa = 10.83APVRR85 pKa = 11.84WKK87 pKa = 11.19LPDD90 pKa = 3.36NYY92 pKa = 10.65LSKK95 pKa = 10.98EE96 pKa = 4.02VFKK99 pKa = 10.68TVLRR103 pKa = 11.84RR104 pKa = 11.84LDD106 pKa = 3.94FSSSPGYY113 pKa = 9.37PYY115 pKa = 10.65CKK117 pKa = 9.28RR118 pKa = 11.84KK119 pKa = 8.01QTIGEE124 pKa = 3.98WLGFDD129 pKa = 4.42GFQFDD134 pKa = 4.22PVQVDD139 pKa = 4.51LLWMDD144 pKa = 3.35VQRR147 pKa = 11.84AILGEE152 pKa = 4.18SEE154 pKa = 4.83LIMRR158 pKa = 11.84VFIKK162 pKa = 10.42QEE164 pKa = 3.47PHH166 pKa = 5.9KK167 pKa = 10.44AAKK170 pKa = 9.93AVEE173 pKa = 4.26GRR175 pKa = 11.84WRR177 pKa = 11.84LIMAFPLAHH186 pKa = 5.15QVLWHH191 pKa = 5.86MLFDD195 pKa = 3.9YY196 pKa = 10.95MNDD199 pKa = 3.52LEE201 pKa = 4.32ITKK204 pKa = 10.21AAEE207 pKa = 3.87IPSQQGIVLYY217 pKa = 10.59GGAWKK222 pKa = 10.25SHH224 pKa = 5.29LARR227 pKa = 11.84WKK229 pKa = 9.3QQGYY233 pKa = 8.17DD234 pKa = 3.13TGLDD238 pKa = 3.15KK239 pKa = 11.3SAWDD243 pKa = 3.15WTYY246 pKa = 10.81TYY248 pKa = 11.07WLMQMDD254 pKa = 4.45LQLRR258 pKa = 11.84YY259 pKa = 8.79RR260 pKa = 11.84TGRR263 pKa = 11.84GFKK266 pKa = 9.85MMEE269 pKa = 3.92WFQLATRR276 pKa = 11.84EE277 pKa = 4.03YY278 pKa = 9.29EE279 pKa = 3.94LAFGVGAKK287 pKa = 10.4FITTDD292 pKa = 3.53GYY294 pKa = 11.06EE295 pKa = 4.15LEE297 pKa = 4.15QLVPGLMKK305 pKa = 10.49SGSVNTISSNSHH317 pKa = 4.99AQAMLHH323 pKa = 6.24ALVCIEE329 pKa = 4.74GGVDD333 pKa = 3.53PDD335 pKa = 5.21PYY337 pKa = 10.23PACCGDD343 pKa = 3.68DD344 pKa = 3.67TLQKK348 pKa = 9.85MSQALIPLYY357 pKa = 9.91EE358 pKa = 4.38KK359 pKa = 10.76YY360 pKa = 10.92GVVVKK365 pKa = 10.55SASEE369 pKa = 3.76GMEE372 pKa = 4.49FIGHH376 pKa = 6.57EE377 pKa = 4.34FLDD380 pKa = 4.22SGPQPLYY387 pKa = 9.82MRR389 pKa = 11.84KK390 pKa = 8.66HH391 pKa = 6.0LNRR394 pKa = 11.84FLHH397 pKa = 6.28MKK399 pKa = 10.45DD400 pKa = 3.94EE401 pKa = 4.18YY402 pKa = 11.38LGDD405 pKa = 3.89YY406 pKa = 10.09LDD408 pKa = 4.2GMSRR412 pKa = 11.84LYY414 pKa = 11.16AKK416 pKa = 9.56TPYY419 pKa = 10.29FEE421 pKa = 4.04FWSWLGDD428 pKa = 3.72VYY430 pKa = 11.26DD431 pKa = 4.14VPVPSKK437 pKa = 10.72QYY439 pKa = 9.19VNNWYY444 pKa = 9.77DD445 pKa = 2.94HH446 pKa = 6.8SYY448 pKa = 11.1
Molecular weight: 51.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KET7|A0A1L3KET7_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 44 OX=1923232 PE=4 SV=1
MM1 pKa = 7.82PCFLGLISEE10 pKa = 4.81LSKK13 pKa = 9.98ITAKK17 pKa = 10.38QQTPPTWKK25 pKa = 10.29DD26 pKa = 3.27FLITPSSLGGSTSSKK41 pKa = 9.59IACTVGATVITVGIGVAVVKK61 pKa = 10.18GWYY64 pKa = 8.62RR65 pKa = 11.84LKK67 pKa = 10.4RR68 pKa = 11.84AVGIRR73 pKa = 11.84PSTKK77 pKa = 10.13VQVTPAAVTNHH88 pKa = 5.91GSRR91 pKa = 11.84EE92 pKa = 3.96SRR94 pKa = 11.84FAGSAEE100 pKa = 4.0VAMPMATGQVLIGFYY115 pKa = 10.7SGDD118 pKa = 3.46EE119 pKa = 4.38YY120 pKa = 10.49ITSGGGYY127 pKa = 10.42RR128 pKa = 11.84MDD130 pKa = 4.18LTTGPYY136 pKa = 10.28LVVPCHH142 pKa = 6.52VLDD145 pKa = 3.78EE146 pKa = 4.56VEE148 pKa = 4.18ARR150 pKa = 11.84GSGEE154 pKa = 3.99VVLRR158 pKa = 11.84GPKK161 pKa = 9.24TDD163 pKa = 3.31ISMKK167 pKa = 10.27LDD169 pKa = 3.43RR170 pKa = 11.84EE171 pKa = 4.31NIAEE175 pKa = 4.87IITDD179 pKa = 3.66VYY181 pKa = 10.28VMKK184 pKa = 10.63VQEE187 pKa = 4.23NLFPNLGVKK196 pKa = 9.63KK197 pKa = 9.79PKK199 pKa = 9.52IGRR202 pKa = 11.84IEE204 pKa = 3.83RR205 pKa = 11.84TASAKK210 pKa = 10.09IAGPGGKK217 pKa = 8.2GTLGDD222 pKa = 3.34ISLQIGQVGMTYY234 pKa = 10.69YY235 pKa = 10.76RR236 pKa = 11.84GTTLPGYY243 pKa = 9.48SGCPYY248 pKa = 10.32QVDD251 pKa = 3.96GNIVGMHH258 pKa = 6.02LRR260 pKa = 11.84GSIVPDD266 pKa = 3.86GPNIGIAARR275 pKa = 11.84MIYY278 pKa = 8.4ITANYY283 pKa = 9.87LLAKK287 pKa = 10.33PEE289 pKa = 5.22DD290 pKa = 3.79SLEE293 pKa = 3.98WLTDD297 pKa = 3.53QVIKK301 pKa = 10.66RR302 pKa = 11.84GNKK305 pKa = 9.71LKK307 pKa = 10.06IDD309 pKa = 4.11KK310 pKa = 10.41NWGSLDD316 pKa = 3.37SYY318 pKa = 10.91RR319 pKa = 11.84FQLGGDD325 pKa = 3.91FAIVEE330 pKa = 4.2SDD332 pKa = 3.76TIYY335 pKa = 10.95EE336 pKa = 4.11ALGEE340 pKa = 4.19DD341 pKa = 3.48WLDD344 pKa = 3.4KK345 pKa = 9.1YY346 pKa = 9.38TEE348 pKa = 4.04YY349 pKa = 11.48SDD351 pKa = 5.35FEE353 pKa = 4.39DD354 pKa = 5.04RR355 pKa = 11.84YY356 pKa = 10.34HH357 pKa = 6.65SKK359 pKa = 10.88KK360 pKa = 9.95KK361 pKa = 10.07GKK363 pKa = 10.08SRR365 pKa = 11.84GEE367 pKa = 3.98SAGNEE372 pKa = 3.92QSPGVSGLPEE382 pKa = 4.19SAQGSQSLVGQQSPPSEE399 pKa = 3.99WKK401 pKa = 10.92LSDD404 pKa = 4.0GLDD407 pKa = 3.26KK408 pKa = 11.13LSRR411 pKa = 11.84KK412 pKa = 8.19QRR414 pKa = 11.84QKK416 pKa = 11.34LSTALKK422 pKa = 10.14AFSAQSQTAA431 pKa = 3.0
MM1 pKa = 7.82PCFLGLISEE10 pKa = 4.81LSKK13 pKa = 9.98ITAKK17 pKa = 10.38QQTPPTWKK25 pKa = 10.29DD26 pKa = 3.27FLITPSSLGGSTSSKK41 pKa = 9.59IACTVGATVITVGIGVAVVKK61 pKa = 10.18GWYY64 pKa = 8.62RR65 pKa = 11.84LKK67 pKa = 10.4RR68 pKa = 11.84AVGIRR73 pKa = 11.84PSTKK77 pKa = 10.13VQVTPAAVTNHH88 pKa = 5.91GSRR91 pKa = 11.84EE92 pKa = 3.96SRR94 pKa = 11.84FAGSAEE100 pKa = 4.0VAMPMATGQVLIGFYY115 pKa = 10.7SGDD118 pKa = 3.46EE119 pKa = 4.38YY120 pKa = 10.49ITSGGGYY127 pKa = 10.42RR128 pKa = 11.84MDD130 pKa = 4.18LTTGPYY136 pKa = 10.28LVVPCHH142 pKa = 6.52VLDD145 pKa = 3.78EE146 pKa = 4.56VEE148 pKa = 4.18ARR150 pKa = 11.84GSGEE154 pKa = 3.99VVLRR158 pKa = 11.84GPKK161 pKa = 9.24TDD163 pKa = 3.31ISMKK167 pKa = 10.27LDD169 pKa = 3.43RR170 pKa = 11.84EE171 pKa = 4.31NIAEE175 pKa = 4.87IITDD179 pKa = 3.66VYY181 pKa = 10.28VMKK184 pKa = 10.63VQEE187 pKa = 4.23NLFPNLGVKK196 pKa = 9.63KK197 pKa = 9.79PKK199 pKa = 9.52IGRR202 pKa = 11.84IEE204 pKa = 3.83RR205 pKa = 11.84TASAKK210 pKa = 10.09IAGPGGKK217 pKa = 8.2GTLGDD222 pKa = 3.34ISLQIGQVGMTYY234 pKa = 10.69YY235 pKa = 10.76RR236 pKa = 11.84GTTLPGYY243 pKa = 9.48SGCPYY248 pKa = 10.32QVDD251 pKa = 3.96GNIVGMHH258 pKa = 6.02LRR260 pKa = 11.84GSIVPDD266 pKa = 3.86GPNIGIAARR275 pKa = 11.84MIYY278 pKa = 8.4ITANYY283 pKa = 9.87LLAKK287 pKa = 10.33PEE289 pKa = 5.22DD290 pKa = 3.79SLEE293 pKa = 3.98WLTDD297 pKa = 3.53QVIKK301 pKa = 10.66RR302 pKa = 11.84GNKK305 pKa = 9.71LKK307 pKa = 10.06IDD309 pKa = 4.11KK310 pKa = 10.41NWGSLDD316 pKa = 3.37SYY318 pKa = 10.91RR319 pKa = 11.84FQLGGDD325 pKa = 3.91FAIVEE330 pKa = 4.2SDD332 pKa = 3.76TIYY335 pKa = 10.95EE336 pKa = 4.11ALGEE340 pKa = 4.19DD341 pKa = 3.48WLDD344 pKa = 3.4KK345 pKa = 9.1YY346 pKa = 9.38TEE348 pKa = 4.04YY349 pKa = 11.48SDD351 pKa = 5.35FEE353 pKa = 4.39DD354 pKa = 5.04RR355 pKa = 11.84YY356 pKa = 10.34HH357 pKa = 6.65SKK359 pKa = 10.88KK360 pKa = 9.95KK361 pKa = 10.07GKK363 pKa = 10.08SRR365 pKa = 11.84GEE367 pKa = 3.98SAGNEE372 pKa = 3.92QSPGVSGLPEE382 pKa = 4.19SAQGSQSLVGQQSPPSEE399 pKa = 3.99WKK401 pKa = 10.92LSDD404 pKa = 4.0GLDD407 pKa = 3.26KK408 pKa = 11.13LSRR411 pKa = 11.84KK412 pKa = 8.19QRR414 pKa = 11.84QKK416 pKa = 11.34LSTALKK422 pKa = 10.14AFSAQSQTAA431 pKa = 3.0
Molecular weight: 46.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
879 |
431 |
448 |
439.5 |
49.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.485 ± 0.008 | 1.138 ± 0.142 |
5.802 ± 0.474 | 5.916 ± 0.551 |
3.299 ± 0.823 | 9.215 ± 1.62 |
1.82 ± 0.606 | 5.006 ± 1.013 |
6.257 ± 0.478 | 9.329 ± 0.978 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.958 ± 0.591 | 2.389 ± 0.047 |
4.778 ± 0.222 | 4.664 ± 0.174 |
4.664 ± 0.016 | 7.281 ± 1.201 |
5.461 ± 0.703 | 6.257 ± 0.478 |
2.503 ± 0.754 | 4.778 ± 0.566 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
