
Hubei narna-like virus 23
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
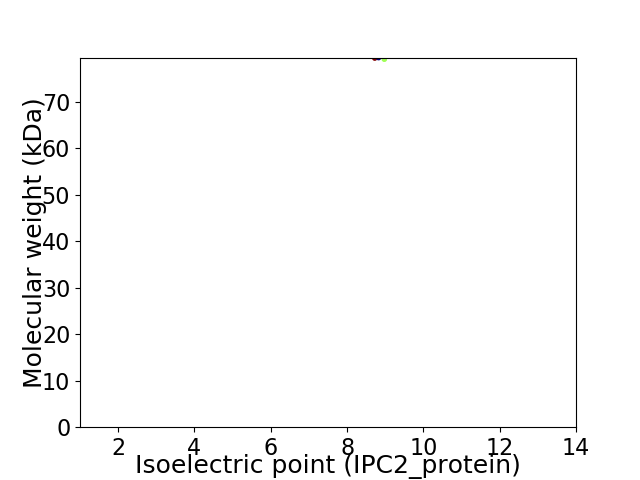
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
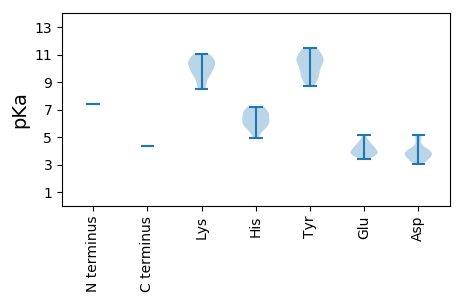
>tr|A0A1L3KIF8|A0A1L3KIF8_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 23 OX=1922954 PE=4 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84KK4 pKa = 8.9NVQRR8 pKa = 11.84GSLCKK13 pKa = 10.09LASPCLGSSTPRR25 pKa = 11.84YY26 pKa = 9.26LHH28 pKa = 6.79QLDD31 pKa = 3.95HH32 pKa = 7.01WIKK35 pKa = 10.08HH36 pKa = 6.05HH37 pKa = 6.1GEE39 pKa = 3.5EE40 pKa = 4.62WTCSRR45 pKa = 11.84LKK47 pKa = 10.18TIANAAMQLRR57 pKa = 11.84SGHH60 pKa = 7.15PDD62 pKa = 3.02TARR65 pKa = 11.84EE66 pKa = 3.75ILEE69 pKa = 4.02EE70 pKa = 3.85GRR72 pKa = 11.84IAVRR76 pKa = 11.84KK77 pKa = 9.49DD78 pKa = 3.0KK79 pKa = 10.43PVPRR83 pKa = 11.84GIMGIVALQYY93 pKa = 11.43VNAQKK98 pKa = 10.54PYY100 pKa = 9.73AVRR103 pKa = 11.84TAACLLRR110 pKa = 11.84AYY112 pKa = 10.6SGIYY116 pKa = 9.36LDD118 pKa = 4.07EE119 pKa = 4.65VSVEE123 pKa = 4.05QVRR126 pKa = 11.84KK127 pKa = 9.57ARR129 pKa = 11.84HH130 pKa = 5.75SICDD134 pKa = 3.58KK135 pKa = 10.85AGKK138 pKa = 9.47GQLLPTHH145 pKa = 7.18PGLQWRR151 pKa = 11.84LIHH154 pKa = 6.75EE155 pKa = 4.48GTKK158 pKa = 9.57MSGLVRR164 pKa = 11.84EE165 pKa = 4.75TPRR168 pKa = 11.84SPVDD172 pKa = 3.55LSYY175 pKa = 11.45LNPSRR180 pKa = 11.84KK181 pKa = 9.39YY182 pKa = 9.2FCGQFRR188 pKa = 11.84LAPEE192 pKa = 5.04LKK194 pKa = 10.24DD195 pKa = 3.57KK196 pKa = 10.39PYY198 pKa = 11.23SSAALSLMTMGSVPKK213 pKa = 10.47DD214 pKa = 4.06LITLCGDD221 pKa = 3.95FALRR225 pKa = 11.84RR226 pKa = 11.84QAEE229 pKa = 4.25RR230 pKa = 11.84FQEE233 pKa = 4.21DD234 pKa = 3.33RR235 pKa = 11.84MNYY238 pKa = 9.62PGFLGKK244 pKa = 9.88ISFLQEE250 pKa = 3.95GGCKK254 pKa = 9.76SRR256 pKa = 11.84TVCQPTAWLQAYY268 pKa = 9.12SMPLAAILCRR278 pKa = 11.84AIYY281 pKa = 10.35LLEE284 pKa = 4.19TGKK287 pKa = 10.97GNRR290 pKa = 11.84RR291 pKa = 11.84SLGLSVVHH299 pKa = 6.49DD300 pKa = 3.95QEE302 pKa = 4.51TGSLFMKK309 pKa = 10.4SAMEE313 pKa = 3.7EE314 pKa = 3.79RR315 pKa = 11.84KK316 pKa = 10.34EE317 pKa = 3.89IFSYY321 pKa = 10.6DD322 pKa = 3.53LKK324 pKa = 11.07SATDD328 pKa = 3.76RR329 pKa = 11.84FPRR332 pKa = 11.84GPQLLLLKK340 pKa = 10.77EE341 pKa = 4.14MGLGDD346 pKa = 3.07WCGAVEE352 pKa = 4.35RR353 pKa = 11.84TAEE356 pKa = 4.12GPYY359 pKa = 8.68WAPEE363 pKa = 3.51ARR365 pKa = 11.84QPWSYY370 pKa = 10.05TVGQPMGLAYY380 pKa = 10.58SFSLFHH386 pKa = 6.0LTHH389 pKa = 6.54RR390 pKa = 11.84AILEE394 pKa = 4.2GLAKK398 pKa = 9.91EE399 pKa = 4.21YY400 pKa = 11.0SPNSPAYY407 pKa = 9.78AVLGDD412 pKa = 4.98DD413 pKa = 4.95VLITCKK419 pKa = 10.48SLALAYY425 pKa = 10.19EE426 pKa = 4.25DD427 pKa = 4.62MISGLGVEE435 pKa = 4.57ISTAKK440 pKa = 10.57SFEE443 pKa = 4.3GPDD446 pKa = 3.5LQSFAGFTGISSKK459 pKa = 10.87KK460 pKa = 9.4GITVFRR466 pKa = 11.84PFKK469 pKa = 10.35HH470 pKa = 6.73GPDD473 pKa = 3.62FQIKK477 pKa = 9.51GKK479 pKa = 8.97EE480 pKa = 3.96LNVLHH485 pKa = 6.25TLGPKK490 pKa = 8.75VRR492 pKa = 11.84EE493 pKa = 3.95WSSWWSKK500 pKa = 10.15QFDD503 pKa = 3.73AYY505 pKa = 10.9RR506 pKa = 11.84CTITFRR512 pKa = 11.84GNDD515 pKa = 4.01LSPLLPEE522 pKa = 4.88DD523 pKa = 3.76QPKK526 pKa = 10.55VGPNRR531 pKa = 11.84PSLGWIGSMVNQLTSPGALRR551 pKa = 11.84IKK553 pKa = 10.84GMGGRR558 pKa = 11.84GCVDD562 pKa = 3.18YY563 pKa = 10.65PVDD566 pKa = 3.99YY567 pKa = 10.58FRR569 pKa = 11.84QEE571 pKa = 3.7HH572 pKa = 5.75NRR574 pKa = 11.84LFLGRR579 pKa = 11.84LSNWFNQKK587 pKa = 8.67AWNHH591 pKa = 5.41EE592 pKa = 3.75FSILVSDD599 pKa = 5.13KK600 pKa = 10.77EE601 pKa = 4.42PNSPAIFAPEE611 pKa = 3.72LLKK614 pKa = 10.9RR615 pKa = 11.84EE616 pKa = 4.58DD617 pKa = 3.98GPFVRR622 pKa = 11.84PYY624 pKa = 10.63SALSTDD630 pKa = 4.1PLIQGHH636 pKa = 5.84RR637 pKa = 11.84HH638 pKa = 4.94DD639 pKa = 3.53QYY641 pKa = 11.39LRR643 pKa = 11.84KK644 pKa = 9.72LRR646 pKa = 11.84EE647 pKa = 3.73EE648 pKa = 3.9SRR650 pKa = 11.84EE651 pKa = 4.15SIRR654 pKa = 11.84KK655 pKa = 8.49QVEE658 pKa = 3.62QEE660 pKa = 3.55ALIRR664 pKa = 11.84AKK666 pKa = 9.92EE667 pKa = 3.86EE668 pKa = 3.55KK669 pKa = 10.01LARR672 pKa = 11.84EE673 pKa = 3.81EE674 pKa = 5.12LEE676 pKa = 3.91RR677 pKa = 11.84QGCLEE682 pKa = 3.8RR683 pKa = 11.84EE684 pKa = 3.4RR685 pKa = 11.84RR686 pKa = 11.84ARR688 pKa = 11.84IKK690 pKa = 11.05APVTKK695 pKa = 8.76PTSRR699 pKa = 11.84STGLGRR705 pKa = 4.33
MM1 pKa = 7.42ARR3 pKa = 11.84KK4 pKa = 8.9NVQRR8 pKa = 11.84GSLCKK13 pKa = 10.09LASPCLGSSTPRR25 pKa = 11.84YY26 pKa = 9.26LHH28 pKa = 6.79QLDD31 pKa = 3.95HH32 pKa = 7.01WIKK35 pKa = 10.08HH36 pKa = 6.05HH37 pKa = 6.1GEE39 pKa = 3.5EE40 pKa = 4.62WTCSRR45 pKa = 11.84LKK47 pKa = 10.18TIANAAMQLRR57 pKa = 11.84SGHH60 pKa = 7.15PDD62 pKa = 3.02TARR65 pKa = 11.84EE66 pKa = 3.75ILEE69 pKa = 4.02EE70 pKa = 3.85GRR72 pKa = 11.84IAVRR76 pKa = 11.84KK77 pKa = 9.49DD78 pKa = 3.0KK79 pKa = 10.43PVPRR83 pKa = 11.84GIMGIVALQYY93 pKa = 11.43VNAQKK98 pKa = 10.54PYY100 pKa = 9.73AVRR103 pKa = 11.84TAACLLRR110 pKa = 11.84AYY112 pKa = 10.6SGIYY116 pKa = 9.36LDD118 pKa = 4.07EE119 pKa = 4.65VSVEE123 pKa = 4.05QVRR126 pKa = 11.84KK127 pKa = 9.57ARR129 pKa = 11.84HH130 pKa = 5.75SICDD134 pKa = 3.58KK135 pKa = 10.85AGKK138 pKa = 9.47GQLLPTHH145 pKa = 7.18PGLQWRR151 pKa = 11.84LIHH154 pKa = 6.75EE155 pKa = 4.48GTKK158 pKa = 9.57MSGLVRR164 pKa = 11.84EE165 pKa = 4.75TPRR168 pKa = 11.84SPVDD172 pKa = 3.55LSYY175 pKa = 11.45LNPSRR180 pKa = 11.84KK181 pKa = 9.39YY182 pKa = 9.2FCGQFRR188 pKa = 11.84LAPEE192 pKa = 5.04LKK194 pKa = 10.24DD195 pKa = 3.57KK196 pKa = 10.39PYY198 pKa = 11.23SSAALSLMTMGSVPKK213 pKa = 10.47DD214 pKa = 4.06LITLCGDD221 pKa = 3.95FALRR225 pKa = 11.84RR226 pKa = 11.84QAEE229 pKa = 4.25RR230 pKa = 11.84FQEE233 pKa = 4.21DD234 pKa = 3.33RR235 pKa = 11.84MNYY238 pKa = 9.62PGFLGKK244 pKa = 9.88ISFLQEE250 pKa = 3.95GGCKK254 pKa = 9.76SRR256 pKa = 11.84TVCQPTAWLQAYY268 pKa = 9.12SMPLAAILCRR278 pKa = 11.84AIYY281 pKa = 10.35LLEE284 pKa = 4.19TGKK287 pKa = 10.97GNRR290 pKa = 11.84RR291 pKa = 11.84SLGLSVVHH299 pKa = 6.49DD300 pKa = 3.95QEE302 pKa = 4.51TGSLFMKK309 pKa = 10.4SAMEE313 pKa = 3.7EE314 pKa = 3.79RR315 pKa = 11.84KK316 pKa = 10.34EE317 pKa = 3.89IFSYY321 pKa = 10.6DD322 pKa = 3.53LKK324 pKa = 11.07SATDD328 pKa = 3.76RR329 pKa = 11.84FPRR332 pKa = 11.84GPQLLLLKK340 pKa = 10.77EE341 pKa = 4.14MGLGDD346 pKa = 3.07WCGAVEE352 pKa = 4.35RR353 pKa = 11.84TAEE356 pKa = 4.12GPYY359 pKa = 8.68WAPEE363 pKa = 3.51ARR365 pKa = 11.84QPWSYY370 pKa = 10.05TVGQPMGLAYY380 pKa = 10.58SFSLFHH386 pKa = 6.0LTHH389 pKa = 6.54RR390 pKa = 11.84AILEE394 pKa = 4.2GLAKK398 pKa = 9.91EE399 pKa = 4.21YY400 pKa = 11.0SPNSPAYY407 pKa = 9.78AVLGDD412 pKa = 4.98DD413 pKa = 4.95VLITCKK419 pKa = 10.48SLALAYY425 pKa = 10.19EE426 pKa = 4.25DD427 pKa = 4.62MISGLGVEE435 pKa = 4.57ISTAKK440 pKa = 10.57SFEE443 pKa = 4.3GPDD446 pKa = 3.5LQSFAGFTGISSKK459 pKa = 10.87KK460 pKa = 9.4GITVFRR466 pKa = 11.84PFKK469 pKa = 10.35HH470 pKa = 6.73GPDD473 pKa = 3.62FQIKK477 pKa = 9.51GKK479 pKa = 8.97EE480 pKa = 3.96LNVLHH485 pKa = 6.25TLGPKK490 pKa = 8.75VRR492 pKa = 11.84EE493 pKa = 3.95WSSWWSKK500 pKa = 10.15QFDD503 pKa = 3.73AYY505 pKa = 10.9RR506 pKa = 11.84CTITFRR512 pKa = 11.84GNDD515 pKa = 4.01LSPLLPEE522 pKa = 4.88DD523 pKa = 3.76QPKK526 pKa = 10.55VGPNRR531 pKa = 11.84PSLGWIGSMVNQLTSPGALRR551 pKa = 11.84IKK553 pKa = 10.84GMGGRR558 pKa = 11.84GCVDD562 pKa = 3.18YY563 pKa = 10.65PVDD566 pKa = 3.99YY567 pKa = 10.58FRR569 pKa = 11.84QEE571 pKa = 3.7HH572 pKa = 5.75NRR574 pKa = 11.84LFLGRR579 pKa = 11.84LSNWFNQKK587 pKa = 8.67AWNHH591 pKa = 5.41EE592 pKa = 3.75FSILVSDD599 pKa = 5.13KK600 pKa = 10.77EE601 pKa = 4.42PNSPAIFAPEE611 pKa = 3.72LLKK614 pKa = 10.9RR615 pKa = 11.84EE616 pKa = 4.58DD617 pKa = 3.98GPFVRR622 pKa = 11.84PYY624 pKa = 10.63SALSTDD630 pKa = 4.1PLIQGHH636 pKa = 5.84RR637 pKa = 11.84HH638 pKa = 4.94DD639 pKa = 3.53QYY641 pKa = 11.39LRR643 pKa = 11.84KK644 pKa = 9.72LRR646 pKa = 11.84EE647 pKa = 3.73EE648 pKa = 3.9SRR650 pKa = 11.84EE651 pKa = 4.15SIRR654 pKa = 11.84KK655 pKa = 8.49QVEE658 pKa = 3.62QEE660 pKa = 3.55ALIRR664 pKa = 11.84AKK666 pKa = 9.92EE667 pKa = 3.86EE668 pKa = 3.55KK669 pKa = 10.01LARR672 pKa = 11.84EE673 pKa = 3.81EE674 pKa = 5.12LEE676 pKa = 3.91RR677 pKa = 11.84QGCLEE682 pKa = 3.8RR683 pKa = 11.84EE684 pKa = 3.4RR685 pKa = 11.84RR686 pKa = 11.84ARR688 pKa = 11.84IKK690 pKa = 11.05APVTKK695 pKa = 8.76PTSRR699 pKa = 11.84STGLGRR705 pKa = 4.33
Molecular weight: 79.4 kDa
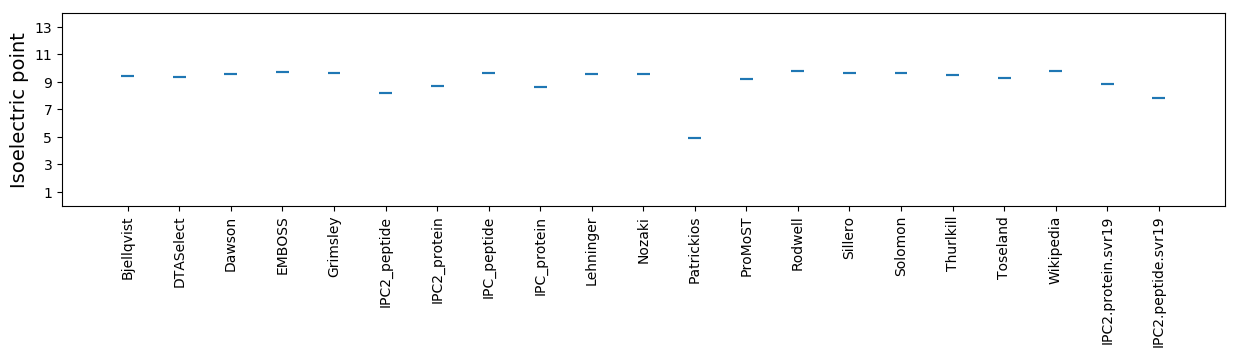
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIF8|A0A1L3KIF8_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 23 OX=1922954 PE=4 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84KK4 pKa = 8.9NVQRR8 pKa = 11.84GSLCKK13 pKa = 10.09LASPCLGSSTPRR25 pKa = 11.84YY26 pKa = 9.26LHH28 pKa = 6.79QLDD31 pKa = 3.95HH32 pKa = 7.01WIKK35 pKa = 10.08HH36 pKa = 6.05HH37 pKa = 6.1GEE39 pKa = 3.5EE40 pKa = 4.62WTCSRR45 pKa = 11.84LKK47 pKa = 10.18TIANAAMQLRR57 pKa = 11.84SGHH60 pKa = 7.15PDD62 pKa = 3.02TARR65 pKa = 11.84EE66 pKa = 3.75ILEE69 pKa = 4.02EE70 pKa = 3.85GRR72 pKa = 11.84IAVRR76 pKa = 11.84KK77 pKa = 9.49DD78 pKa = 3.0KK79 pKa = 10.43PVPRR83 pKa = 11.84GIMGIVALQYY93 pKa = 11.43VNAQKK98 pKa = 10.54PYY100 pKa = 9.73AVRR103 pKa = 11.84TAACLLRR110 pKa = 11.84AYY112 pKa = 10.6SGIYY116 pKa = 9.36LDD118 pKa = 4.07EE119 pKa = 4.65VSVEE123 pKa = 4.05QVRR126 pKa = 11.84KK127 pKa = 9.57ARR129 pKa = 11.84HH130 pKa = 5.75SICDD134 pKa = 3.58KK135 pKa = 10.85AGKK138 pKa = 9.47GQLLPTHH145 pKa = 7.18PGLQWRR151 pKa = 11.84LIHH154 pKa = 6.75EE155 pKa = 4.48GTKK158 pKa = 9.57MSGLVRR164 pKa = 11.84EE165 pKa = 4.75TPRR168 pKa = 11.84SPVDD172 pKa = 3.55LSYY175 pKa = 11.45LNPSRR180 pKa = 11.84KK181 pKa = 9.39YY182 pKa = 9.2FCGQFRR188 pKa = 11.84LAPEE192 pKa = 5.04LKK194 pKa = 10.24DD195 pKa = 3.57KK196 pKa = 10.39PYY198 pKa = 11.23SSAALSLMTMGSVPKK213 pKa = 10.47DD214 pKa = 4.06LITLCGDD221 pKa = 3.95FALRR225 pKa = 11.84RR226 pKa = 11.84QAEE229 pKa = 4.25RR230 pKa = 11.84FQEE233 pKa = 4.21DD234 pKa = 3.33RR235 pKa = 11.84MNYY238 pKa = 9.62PGFLGKK244 pKa = 9.88ISFLQEE250 pKa = 3.95GGCKK254 pKa = 9.76SRR256 pKa = 11.84TVCQPTAWLQAYY268 pKa = 9.12SMPLAAILCRR278 pKa = 11.84AIYY281 pKa = 10.35LLEE284 pKa = 4.19TGKK287 pKa = 10.97GNRR290 pKa = 11.84RR291 pKa = 11.84SLGLSVVHH299 pKa = 6.49DD300 pKa = 3.95QEE302 pKa = 4.51TGSLFMKK309 pKa = 10.4SAMEE313 pKa = 3.7EE314 pKa = 3.79RR315 pKa = 11.84KK316 pKa = 10.34EE317 pKa = 3.89IFSYY321 pKa = 10.6DD322 pKa = 3.53LKK324 pKa = 11.07SATDD328 pKa = 3.76RR329 pKa = 11.84FPRR332 pKa = 11.84GPQLLLLKK340 pKa = 10.77EE341 pKa = 4.14MGLGDD346 pKa = 3.07WCGAVEE352 pKa = 4.35RR353 pKa = 11.84TAEE356 pKa = 4.12GPYY359 pKa = 8.68WAPEE363 pKa = 3.51ARR365 pKa = 11.84QPWSYY370 pKa = 10.05TVGQPMGLAYY380 pKa = 10.58SFSLFHH386 pKa = 6.0LTHH389 pKa = 6.54RR390 pKa = 11.84AILEE394 pKa = 4.2GLAKK398 pKa = 9.91EE399 pKa = 4.21YY400 pKa = 11.0SPNSPAYY407 pKa = 9.78AVLGDD412 pKa = 4.98DD413 pKa = 4.95VLITCKK419 pKa = 10.48SLALAYY425 pKa = 10.19EE426 pKa = 4.25DD427 pKa = 4.62MISGLGVEE435 pKa = 4.57ISTAKK440 pKa = 10.57SFEE443 pKa = 4.3GPDD446 pKa = 3.5LQSFAGFTGISSKK459 pKa = 10.87KK460 pKa = 9.4GITVFRR466 pKa = 11.84PFKK469 pKa = 10.35HH470 pKa = 6.73GPDD473 pKa = 3.62FQIKK477 pKa = 9.51GKK479 pKa = 8.97EE480 pKa = 3.96LNVLHH485 pKa = 6.25TLGPKK490 pKa = 8.75VRR492 pKa = 11.84EE493 pKa = 3.95WSSWWSKK500 pKa = 10.15QFDD503 pKa = 3.73AYY505 pKa = 10.9RR506 pKa = 11.84CTITFRR512 pKa = 11.84GNDD515 pKa = 4.01LSPLLPEE522 pKa = 4.88DD523 pKa = 3.76QPKK526 pKa = 10.55VGPNRR531 pKa = 11.84PSLGWIGSMVNQLTSPGALRR551 pKa = 11.84IKK553 pKa = 10.84GMGGRR558 pKa = 11.84GCVDD562 pKa = 3.18YY563 pKa = 10.65PVDD566 pKa = 3.99YY567 pKa = 10.58FRR569 pKa = 11.84QEE571 pKa = 3.7HH572 pKa = 5.75NRR574 pKa = 11.84LFLGRR579 pKa = 11.84LSNWFNQKK587 pKa = 8.67AWNHH591 pKa = 5.41EE592 pKa = 3.75FSILVSDD599 pKa = 5.13KK600 pKa = 10.77EE601 pKa = 4.42PNSPAIFAPEE611 pKa = 3.72LLKK614 pKa = 10.9RR615 pKa = 11.84EE616 pKa = 4.58DD617 pKa = 3.98GPFVRR622 pKa = 11.84PYY624 pKa = 10.63SALSTDD630 pKa = 4.1PLIQGHH636 pKa = 5.84RR637 pKa = 11.84HH638 pKa = 4.94DD639 pKa = 3.53QYY641 pKa = 11.39LRR643 pKa = 11.84KK644 pKa = 9.72LRR646 pKa = 11.84EE647 pKa = 3.73EE648 pKa = 3.9SRR650 pKa = 11.84EE651 pKa = 4.15SIRR654 pKa = 11.84KK655 pKa = 8.49QVEE658 pKa = 3.62QEE660 pKa = 3.55ALIRR664 pKa = 11.84AKK666 pKa = 9.92EE667 pKa = 3.86EE668 pKa = 3.55KK669 pKa = 10.01LARR672 pKa = 11.84EE673 pKa = 3.81EE674 pKa = 5.12LEE676 pKa = 3.91RR677 pKa = 11.84QGCLEE682 pKa = 3.8RR683 pKa = 11.84EE684 pKa = 3.4RR685 pKa = 11.84RR686 pKa = 11.84ARR688 pKa = 11.84IKK690 pKa = 11.05APVTKK695 pKa = 8.76PTSRR699 pKa = 11.84STGLGRR705 pKa = 4.33
MM1 pKa = 7.42ARR3 pKa = 11.84KK4 pKa = 8.9NVQRR8 pKa = 11.84GSLCKK13 pKa = 10.09LASPCLGSSTPRR25 pKa = 11.84YY26 pKa = 9.26LHH28 pKa = 6.79QLDD31 pKa = 3.95HH32 pKa = 7.01WIKK35 pKa = 10.08HH36 pKa = 6.05HH37 pKa = 6.1GEE39 pKa = 3.5EE40 pKa = 4.62WTCSRR45 pKa = 11.84LKK47 pKa = 10.18TIANAAMQLRR57 pKa = 11.84SGHH60 pKa = 7.15PDD62 pKa = 3.02TARR65 pKa = 11.84EE66 pKa = 3.75ILEE69 pKa = 4.02EE70 pKa = 3.85GRR72 pKa = 11.84IAVRR76 pKa = 11.84KK77 pKa = 9.49DD78 pKa = 3.0KK79 pKa = 10.43PVPRR83 pKa = 11.84GIMGIVALQYY93 pKa = 11.43VNAQKK98 pKa = 10.54PYY100 pKa = 9.73AVRR103 pKa = 11.84TAACLLRR110 pKa = 11.84AYY112 pKa = 10.6SGIYY116 pKa = 9.36LDD118 pKa = 4.07EE119 pKa = 4.65VSVEE123 pKa = 4.05QVRR126 pKa = 11.84KK127 pKa = 9.57ARR129 pKa = 11.84HH130 pKa = 5.75SICDD134 pKa = 3.58KK135 pKa = 10.85AGKK138 pKa = 9.47GQLLPTHH145 pKa = 7.18PGLQWRR151 pKa = 11.84LIHH154 pKa = 6.75EE155 pKa = 4.48GTKK158 pKa = 9.57MSGLVRR164 pKa = 11.84EE165 pKa = 4.75TPRR168 pKa = 11.84SPVDD172 pKa = 3.55LSYY175 pKa = 11.45LNPSRR180 pKa = 11.84KK181 pKa = 9.39YY182 pKa = 9.2FCGQFRR188 pKa = 11.84LAPEE192 pKa = 5.04LKK194 pKa = 10.24DD195 pKa = 3.57KK196 pKa = 10.39PYY198 pKa = 11.23SSAALSLMTMGSVPKK213 pKa = 10.47DD214 pKa = 4.06LITLCGDD221 pKa = 3.95FALRR225 pKa = 11.84RR226 pKa = 11.84QAEE229 pKa = 4.25RR230 pKa = 11.84FQEE233 pKa = 4.21DD234 pKa = 3.33RR235 pKa = 11.84MNYY238 pKa = 9.62PGFLGKK244 pKa = 9.88ISFLQEE250 pKa = 3.95GGCKK254 pKa = 9.76SRR256 pKa = 11.84TVCQPTAWLQAYY268 pKa = 9.12SMPLAAILCRR278 pKa = 11.84AIYY281 pKa = 10.35LLEE284 pKa = 4.19TGKK287 pKa = 10.97GNRR290 pKa = 11.84RR291 pKa = 11.84SLGLSVVHH299 pKa = 6.49DD300 pKa = 3.95QEE302 pKa = 4.51TGSLFMKK309 pKa = 10.4SAMEE313 pKa = 3.7EE314 pKa = 3.79RR315 pKa = 11.84KK316 pKa = 10.34EE317 pKa = 3.89IFSYY321 pKa = 10.6DD322 pKa = 3.53LKK324 pKa = 11.07SATDD328 pKa = 3.76RR329 pKa = 11.84FPRR332 pKa = 11.84GPQLLLLKK340 pKa = 10.77EE341 pKa = 4.14MGLGDD346 pKa = 3.07WCGAVEE352 pKa = 4.35RR353 pKa = 11.84TAEE356 pKa = 4.12GPYY359 pKa = 8.68WAPEE363 pKa = 3.51ARR365 pKa = 11.84QPWSYY370 pKa = 10.05TVGQPMGLAYY380 pKa = 10.58SFSLFHH386 pKa = 6.0LTHH389 pKa = 6.54RR390 pKa = 11.84AILEE394 pKa = 4.2GLAKK398 pKa = 9.91EE399 pKa = 4.21YY400 pKa = 11.0SPNSPAYY407 pKa = 9.78AVLGDD412 pKa = 4.98DD413 pKa = 4.95VLITCKK419 pKa = 10.48SLALAYY425 pKa = 10.19EE426 pKa = 4.25DD427 pKa = 4.62MISGLGVEE435 pKa = 4.57ISTAKK440 pKa = 10.57SFEE443 pKa = 4.3GPDD446 pKa = 3.5LQSFAGFTGISSKK459 pKa = 10.87KK460 pKa = 9.4GITVFRR466 pKa = 11.84PFKK469 pKa = 10.35HH470 pKa = 6.73GPDD473 pKa = 3.62FQIKK477 pKa = 9.51GKK479 pKa = 8.97EE480 pKa = 3.96LNVLHH485 pKa = 6.25TLGPKK490 pKa = 8.75VRR492 pKa = 11.84EE493 pKa = 3.95WSSWWSKK500 pKa = 10.15QFDD503 pKa = 3.73AYY505 pKa = 10.9RR506 pKa = 11.84CTITFRR512 pKa = 11.84GNDD515 pKa = 4.01LSPLLPEE522 pKa = 4.88DD523 pKa = 3.76QPKK526 pKa = 10.55VGPNRR531 pKa = 11.84PSLGWIGSMVNQLTSPGALRR551 pKa = 11.84IKK553 pKa = 10.84GMGGRR558 pKa = 11.84GCVDD562 pKa = 3.18YY563 pKa = 10.65PVDD566 pKa = 3.99YY567 pKa = 10.58FRR569 pKa = 11.84QEE571 pKa = 3.7HH572 pKa = 5.75NRR574 pKa = 11.84LFLGRR579 pKa = 11.84LSNWFNQKK587 pKa = 8.67AWNHH591 pKa = 5.41EE592 pKa = 3.75FSILVSDD599 pKa = 5.13KK600 pKa = 10.77EE601 pKa = 4.42PNSPAIFAPEE611 pKa = 3.72LLKK614 pKa = 10.9RR615 pKa = 11.84EE616 pKa = 4.58DD617 pKa = 3.98GPFVRR622 pKa = 11.84PYY624 pKa = 10.63SALSTDD630 pKa = 4.1PLIQGHH636 pKa = 5.84RR637 pKa = 11.84HH638 pKa = 4.94DD639 pKa = 3.53QYY641 pKa = 11.39LRR643 pKa = 11.84KK644 pKa = 9.72LRR646 pKa = 11.84EE647 pKa = 3.73EE648 pKa = 3.9SRR650 pKa = 11.84EE651 pKa = 4.15SIRR654 pKa = 11.84KK655 pKa = 8.49QVEE658 pKa = 3.62QEE660 pKa = 3.55ALIRR664 pKa = 11.84AKK666 pKa = 9.92EE667 pKa = 3.86EE668 pKa = 3.55KK669 pKa = 10.01LARR672 pKa = 11.84EE673 pKa = 3.81EE674 pKa = 5.12LEE676 pKa = 3.91RR677 pKa = 11.84QGCLEE682 pKa = 3.8RR683 pKa = 11.84EE684 pKa = 3.4RR685 pKa = 11.84RR686 pKa = 11.84ARR688 pKa = 11.84IKK690 pKa = 11.05APVTKK695 pKa = 8.76PTSRR699 pKa = 11.84STGLGRR705 pKa = 4.33
Molecular weight: 79.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
705 |
705 |
705 |
705.0 |
79.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.234 ± 0.0 | 2.128 ± 0.0 |
3.972 ± 0.0 | 6.667 ± 0.0 |
3.546 ± 0.0 | 8.085 ± 0.0 |
2.411 ± 0.0 | 4.255 ± 0.0 |
6.099 ± 0.0 | 10.922 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.128 ± 0.0 | 2.27 ± 0.0 |
6.241 ± 0.0 | 4.255 ± 0.0 |
8.227 ± 0.0 | 7.801 ± 0.0 |
4.255 ± 0.0 | 4.397 ± 0.0 |
1.844 ± 0.0 | 3.262 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
