
Xanthomonas phage Xf409
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Xylivirus; Xanthomonas virus Xf109
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins
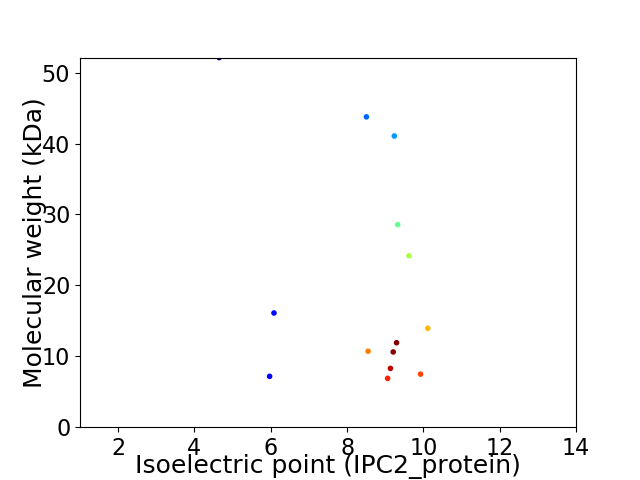
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6DXV6|A0A1W6DXV6_9VIRU Uncharacterized protein OS=Xanthomonas phage Xf409 OX=1979540 PE=4 SV=1
MM1 pKa = 7.94GIGQGSGDD9 pKa = 3.41GRR11 pKa = 11.84VDD13 pKa = 3.68LVGRR17 pKa = 11.84LAGGLRR23 pKa = 11.84DD24 pKa = 3.7CLRR27 pKa = 11.84GFRR30 pKa = 11.84IMGWLARR37 pKa = 11.84VFASAIARR45 pKa = 11.84RR46 pKa = 11.84LAYY49 pKa = 10.38VLVAATLAWCGIGCASAEE67 pKa = 4.1DD68 pKa = 4.89FGTQGAAYY76 pKa = 8.25SACMSQTAAYY86 pKa = 8.6LASRR90 pKa = 11.84GRR92 pKa = 11.84GANDD96 pKa = 3.61RR97 pKa = 11.84NPEE100 pKa = 4.17CNVEE104 pKa = 4.26QGSHH108 pKa = 6.48AYY110 pKa = 9.94RR111 pKa = 11.84GQFEE115 pKa = 4.32TRR117 pKa = 11.84DD118 pKa = 3.73CSSCDD123 pKa = 2.74WYY125 pKa = 10.62RR126 pKa = 11.84AYY128 pKa = 10.79YY129 pKa = 10.43GYY131 pKa = 10.27FAWSKK136 pKa = 11.32GCDD139 pKa = 3.39QEE141 pKa = 4.35PDD143 pKa = 3.69YY144 pKa = 11.04TGSGPWGTYY153 pKa = 9.79VGTARR158 pKa = 11.84NGSIGCRR165 pKa = 11.84NGCDD169 pKa = 3.53GVWFGNSDD177 pKa = 4.83DD178 pKa = 3.88TMTWNATGAICPKK191 pKa = 10.5DD192 pKa = 3.59PEE194 pKa = 4.47KK195 pKa = 10.66TCDD198 pKa = 3.33AMTGGYY204 pKa = 8.06VWNGYY209 pKa = 10.15LGVCEE214 pKa = 4.54PPPNEE219 pKa = 4.34KK220 pKa = 10.53CPEE223 pKa = 4.08GQVPDD228 pKa = 4.2GKK230 pKa = 10.9GGCSPNKK237 pKa = 9.93CPEE240 pKa = 3.85GMLLQADD247 pKa = 4.98GTCAPKK253 pKa = 10.83KK254 pKa = 9.58NDD256 pKa = 3.67CPAGQIRR263 pKa = 11.84SPSGSCLPGDD273 pKa = 4.19GQCAAGEE280 pKa = 4.2VRR282 pKa = 11.84GPDD285 pKa = 3.62GTCKK289 pKa = 10.24KK290 pKa = 10.69DD291 pKa = 3.18GDD293 pKa = 4.3GDD295 pKa = 4.23GEE297 pKa = 4.16PDD299 pKa = 3.55EE300 pKa = 5.35PGEE303 pKa = 4.14GDD305 pKa = 2.97KK306 pKa = 11.39SQFSGGDD313 pKa = 3.37SCDD316 pKa = 3.89SPPSCSGDD324 pKa = 3.87AIMCGQARR332 pKa = 11.84IQWRR336 pKa = 11.84IDD338 pKa = 3.14CNTRR342 pKa = 11.84RR343 pKa = 11.84DD344 pKa = 3.65VNITGGSCAAMPVCVGTNCKK364 pKa = 9.99ALEE367 pKa = 3.92YY368 pKa = 10.61SQLLMQWRR376 pKa = 11.84AACALEE382 pKa = 4.09KK383 pKa = 10.52AANNSGGGTGNNADD397 pKa = 3.25VKK399 pKa = 10.58AIRR402 pKa = 11.84DD403 pKa = 4.11AITGNGTADD412 pKa = 2.97IGADD416 pKa = 3.86GKK418 pKa = 10.07PADD421 pKa = 4.27AFSDD425 pKa = 3.52EE426 pKa = 4.31SGYY429 pKa = 10.46GQEE432 pKa = 4.84GYY434 pKa = 7.72PTGEE438 pKa = 5.06LDD440 pKa = 3.66TQGFGYY446 pKa = 10.49SRR448 pKa = 11.84TCPTIPDD455 pKa = 3.49VAVFGQTLHH464 pKa = 6.82FDD466 pKa = 3.53TSKK469 pKa = 9.79FCQWMVLGGQIVLVMASLVSLRR491 pKa = 11.84LMSQGGSAA499 pKa = 3.86
MM1 pKa = 7.94GIGQGSGDD9 pKa = 3.41GRR11 pKa = 11.84VDD13 pKa = 3.68LVGRR17 pKa = 11.84LAGGLRR23 pKa = 11.84DD24 pKa = 3.7CLRR27 pKa = 11.84GFRR30 pKa = 11.84IMGWLARR37 pKa = 11.84VFASAIARR45 pKa = 11.84RR46 pKa = 11.84LAYY49 pKa = 10.38VLVAATLAWCGIGCASAEE67 pKa = 4.1DD68 pKa = 4.89FGTQGAAYY76 pKa = 8.25SACMSQTAAYY86 pKa = 8.6LASRR90 pKa = 11.84GRR92 pKa = 11.84GANDD96 pKa = 3.61RR97 pKa = 11.84NPEE100 pKa = 4.17CNVEE104 pKa = 4.26QGSHH108 pKa = 6.48AYY110 pKa = 9.94RR111 pKa = 11.84GQFEE115 pKa = 4.32TRR117 pKa = 11.84DD118 pKa = 3.73CSSCDD123 pKa = 2.74WYY125 pKa = 10.62RR126 pKa = 11.84AYY128 pKa = 10.79YY129 pKa = 10.43GYY131 pKa = 10.27FAWSKK136 pKa = 11.32GCDD139 pKa = 3.39QEE141 pKa = 4.35PDD143 pKa = 3.69YY144 pKa = 11.04TGSGPWGTYY153 pKa = 9.79VGTARR158 pKa = 11.84NGSIGCRR165 pKa = 11.84NGCDD169 pKa = 3.53GVWFGNSDD177 pKa = 4.83DD178 pKa = 3.88TMTWNATGAICPKK191 pKa = 10.5DD192 pKa = 3.59PEE194 pKa = 4.47KK195 pKa = 10.66TCDD198 pKa = 3.33AMTGGYY204 pKa = 8.06VWNGYY209 pKa = 10.15LGVCEE214 pKa = 4.54PPPNEE219 pKa = 4.34KK220 pKa = 10.53CPEE223 pKa = 4.08GQVPDD228 pKa = 4.2GKK230 pKa = 10.9GGCSPNKK237 pKa = 9.93CPEE240 pKa = 3.85GMLLQADD247 pKa = 4.98GTCAPKK253 pKa = 10.83KK254 pKa = 9.58NDD256 pKa = 3.67CPAGQIRR263 pKa = 11.84SPSGSCLPGDD273 pKa = 4.19GQCAAGEE280 pKa = 4.2VRR282 pKa = 11.84GPDD285 pKa = 3.62GTCKK289 pKa = 10.24KK290 pKa = 10.69DD291 pKa = 3.18GDD293 pKa = 4.3GDD295 pKa = 4.23GEE297 pKa = 4.16PDD299 pKa = 3.55EE300 pKa = 5.35PGEE303 pKa = 4.14GDD305 pKa = 2.97KK306 pKa = 11.39SQFSGGDD313 pKa = 3.37SCDD316 pKa = 3.89SPPSCSGDD324 pKa = 3.87AIMCGQARR332 pKa = 11.84IQWRR336 pKa = 11.84IDD338 pKa = 3.14CNTRR342 pKa = 11.84RR343 pKa = 11.84DD344 pKa = 3.65VNITGGSCAAMPVCVGTNCKK364 pKa = 9.99ALEE367 pKa = 3.92YY368 pKa = 10.61SQLLMQWRR376 pKa = 11.84AACALEE382 pKa = 4.09KK383 pKa = 10.52AANNSGGGTGNNADD397 pKa = 3.25VKK399 pKa = 10.58AIRR402 pKa = 11.84DD403 pKa = 4.11AITGNGTADD412 pKa = 2.97IGADD416 pKa = 3.86GKK418 pKa = 10.07PADD421 pKa = 4.27AFSDD425 pKa = 3.52EE426 pKa = 4.31SGYY429 pKa = 10.46GQEE432 pKa = 4.84GYY434 pKa = 7.72PTGEE438 pKa = 5.06LDD440 pKa = 3.66TQGFGYY446 pKa = 10.49SRR448 pKa = 11.84TCPTIPDD455 pKa = 3.49VAVFGQTLHH464 pKa = 6.82FDD466 pKa = 3.53TSKK469 pKa = 9.79FCQWMVLGGQIVLVMASLVSLRR491 pKa = 11.84LMSQGGSAA499 pKa = 3.86
Molecular weight: 52.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6DY99|A0A1W6DY99_9VIRU Uncharacterized protein OS=Xanthomonas phage Xf409 OX=1979540 PE=4 SV=1
MM1 pKa = 7.21SAEE4 pKa = 4.22NEE6 pKa = 4.36LIDD9 pKa = 4.9LVRR12 pKa = 11.84AGGKK16 pKa = 9.71FSSDD20 pKa = 2.66NALAQKK26 pKa = 10.69LGITRR31 pKa = 11.84AMVSSWRR38 pKa = 11.84SGRR41 pKa = 11.84YY42 pKa = 9.32AMPDD46 pKa = 3.28DD47 pKa = 5.94QIAQLCALAKK57 pKa = 10.39LDD59 pKa = 3.74GASWMARR66 pKa = 11.84IHH68 pKa = 5.15TEE70 pKa = 3.65RR71 pKa = 11.84AGSATEE77 pKa = 3.71RR78 pKa = 11.84ALWQSILDD86 pKa = 3.67RR87 pKa = 11.84LTPITAVVGALALCEE102 pKa = 3.95IRR104 pKa = 11.84NRR106 pKa = 11.84AVDD109 pKa = 3.32AALHH113 pKa = 5.65RR114 pKa = 11.84RR115 pKa = 11.84GLLLQPTPQEE125 pKa = 4.41ADD127 pKa = 2.55WTMTFDD133 pKa = 2.91TYY135 pKa = 11.54EE136 pKa = 4.25RR137 pKa = 11.84VDD139 pKa = 4.05LTGPWAGFGFQGHH152 pKa = 6.07RR153 pKa = 11.84FFTPEE158 pKa = 4.2NYY160 pKa = 10.47DD161 pKa = 3.09IDD163 pKa = 4.44PCGMRR168 pKa = 11.84YY169 pKa = 8.0WALTCAIARR178 pKa = 11.84EE179 pKa = 4.08WSLMMSEE186 pKa = 4.18EE187 pKa = 4.07RR188 pKa = 11.84NARR191 pKa = 11.84SANPRR196 pKa = 11.84TPTATRR202 pKa = 11.84SPGSRR207 pKa = 11.84ISADD211 pKa = 3.04ANVIYY216 pKa = 10.62LRR218 pKa = 11.84DD219 pKa = 3.39VLRR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84EE226 pKa = 3.68TRR228 pKa = 11.84LSVGRR233 pKa = 11.84DD234 pKa = 3.15PGLADD239 pKa = 3.28SGKK242 pKa = 10.03RR243 pKa = 11.84SPAGRR248 pKa = 11.84GRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84PRR254 pKa = 11.84RR255 pKa = 11.84GG256 pKa = 2.87
MM1 pKa = 7.21SAEE4 pKa = 4.22NEE6 pKa = 4.36LIDD9 pKa = 4.9LVRR12 pKa = 11.84AGGKK16 pKa = 9.71FSSDD20 pKa = 2.66NALAQKK26 pKa = 10.69LGITRR31 pKa = 11.84AMVSSWRR38 pKa = 11.84SGRR41 pKa = 11.84YY42 pKa = 9.32AMPDD46 pKa = 3.28DD47 pKa = 5.94QIAQLCALAKK57 pKa = 10.39LDD59 pKa = 3.74GASWMARR66 pKa = 11.84IHH68 pKa = 5.15TEE70 pKa = 3.65RR71 pKa = 11.84AGSATEE77 pKa = 3.71RR78 pKa = 11.84ALWQSILDD86 pKa = 3.67RR87 pKa = 11.84LTPITAVVGALALCEE102 pKa = 3.95IRR104 pKa = 11.84NRR106 pKa = 11.84AVDD109 pKa = 3.32AALHH113 pKa = 5.65RR114 pKa = 11.84RR115 pKa = 11.84GLLLQPTPQEE125 pKa = 4.41ADD127 pKa = 2.55WTMTFDD133 pKa = 2.91TYY135 pKa = 11.54EE136 pKa = 4.25RR137 pKa = 11.84VDD139 pKa = 4.05LTGPWAGFGFQGHH152 pKa = 6.07RR153 pKa = 11.84FFTPEE158 pKa = 4.2NYY160 pKa = 10.47DD161 pKa = 3.09IDD163 pKa = 4.44PCGMRR168 pKa = 11.84YY169 pKa = 8.0WALTCAIARR178 pKa = 11.84EE179 pKa = 4.08WSLMMSEE186 pKa = 4.18EE187 pKa = 4.07RR188 pKa = 11.84NARR191 pKa = 11.84SANPRR196 pKa = 11.84TPTATRR202 pKa = 11.84SPGSRR207 pKa = 11.84ISADD211 pKa = 3.04ANVIYY216 pKa = 10.62LRR218 pKa = 11.84DD219 pKa = 3.39VLRR222 pKa = 11.84RR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84EE226 pKa = 3.68TRR228 pKa = 11.84LSVGRR233 pKa = 11.84DD234 pKa = 3.15PGLADD239 pKa = 3.28SGKK242 pKa = 10.03RR243 pKa = 11.84SPAGRR248 pKa = 11.84GRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84PRR254 pKa = 11.84RR255 pKa = 11.84GG256 pKa = 2.87
Molecular weight: 28.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2591 |
61 |
499 |
185.1 |
20.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.35 ± 0.927 | 2.239 ± 0.828 |
5.288 ± 0.599 | 3.782 ± 0.395 |
3.396 ± 0.54 | 9.726 ± 1.281 |
1.621 ± 0.349 | 3.705 ± 0.359 |
3.782 ± 0.601 | 8.452 ± 1.17 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.236 | 2.277 ± 0.378 |
5.21 ± 0.438 | 4.052 ± 0.512 |
8.066 ± 1.096 | 6.059 ± 0.511 |
5.403 ± 0.721 | 6.561 ± 0.994 |
2.74 ± 0.424 | 2.74 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
