
Caenorhabditis japonica
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Rhabditina; Rhabditomorpha; Rhabditoidea; Rhabditidae; Peloderinae; Caenorhabditis
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
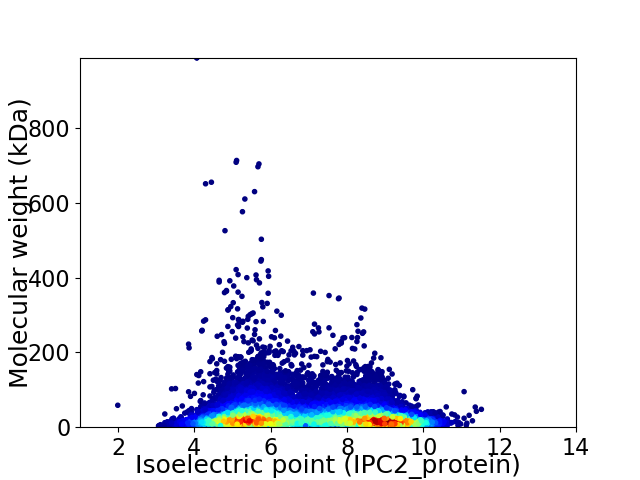
Virtual 2D-PAGE plot for 35026 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H2JLH8|A0A2H2JLH8_CAEJA Uncharacterized protein OS=Caenorhabditis japonica OX=281687 PE=4 SV=1
MM1 pKa = 7.56YY2 pKa = 10.48INDD5 pKa = 4.73LLEE8 pKa = 4.24MFPPDD13 pKa = 2.88IHH15 pKa = 5.67VTAFADD21 pKa = 3.85DD22 pKa = 4.4LKK24 pKa = 11.29LLSSNPASLQHH35 pKa = 6.55SINLVADD42 pKa = 4.13WFSILDD48 pKa = 3.91TANQAAPSYY57 pKa = 8.09TIPSDD62 pKa = 3.47SGPCSSCLEE71 pKa = 4.21YY72 pKa = 11.11ANSSAAVPEE81 pKa = 4.23VAPEE85 pKa = 3.87GLIVDD90 pKa = 4.05EE91 pKa = 5.21DD92 pKa = 4.06FDD94 pKa = 6.55RR95 pKa = 11.84ILDD98 pKa = 3.67VVGPSFRR105 pKa = 11.84RR106 pKa = 11.84TTAWSCC112 pKa = 3.12
MM1 pKa = 7.56YY2 pKa = 10.48INDD5 pKa = 4.73LLEE8 pKa = 4.24MFPPDD13 pKa = 2.88IHH15 pKa = 5.67VTAFADD21 pKa = 3.85DD22 pKa = 4.4LKK24 pKa = 11.29LLSSNPASLQHH35 pKa = 6.55SINLVADD42 pKa = 4.13WFSILDD48 pKa = 3.91TANQAAPSYY57 pKa = 8.09TIPSDD62 pKa = 3.47SGPCSSCLEE71 pKa = 4.21YY72 pKa = 11.11ANSSAAVPEE81 pKa = 4.23VAPEE85 pKa = 3.87GLIVDD90 pKa = 4.05EE91 pKa = 5.21DD92 pKa = 4.06FDD94 pKa = 6.55RR95 pKa = 11.84ILDD98 pKa = 3.67VVGPSFRR105 pKa = 11.84RR106 pKa = 11.84TTAWSCC112 pKa = 3.12
Molecular weight: 12.15 kDa
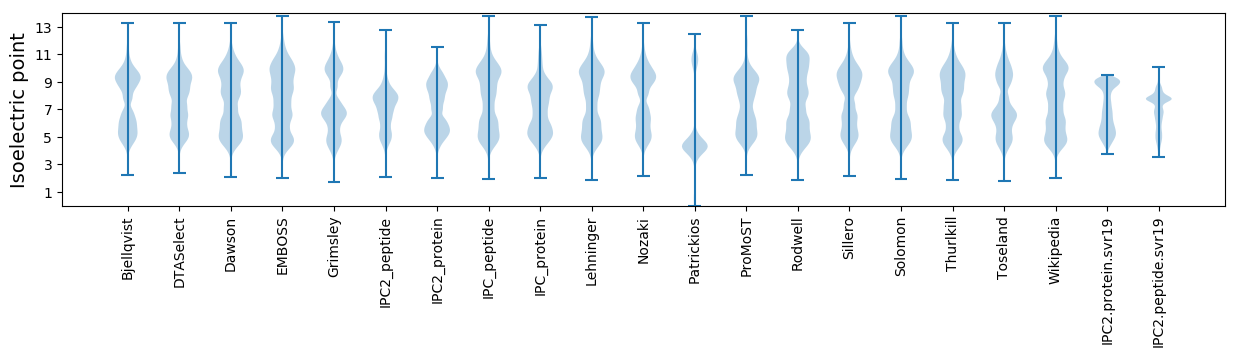
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H2IB26|A0A2H2IB26_CAEJA Uncharacterized protein OS=Caenorhabditis japonica OX=281687 PE=4 SV=1
MM1 pKa = 7.46YY2 pKa = 10.32SRR4 pKa = 11.84SPPPRR9 pKa = 11.84GQLPDD14 pKa = 3.48SYY16 pKa = 11.18LAAVWRR22 pKa = 11.84LLGGSLAALWRR33 pKa = 11.84LFGGSLAALWRR44 pKa = 11.84LCVGYY49 pKa = 8.19RR50 pKa = 11.84TARR53 pKa = 11.84NRR55 pKa = 11.84TARR58 pKa = 11.84NRR60 pKa = 11.84TARR63 pKa = 11.84NRR65 pKa = 11.84TTRR68 pKa = 11.84NRR70 pKa = 11.84TARR73 pKa = 11.84NRR75 pKa = 11.84TTRR78 pKa = 11.84NRR80 pKa = 11.84TARR83 pKa = 11.84SRR85 pKa = 11.84TARR88 pKa = 11.84SRR90 pKa = 11.84TARR93 pKa = 11.84NRR95 pKa = 11.84TARR98 pKa = 11.84SRR100 pKa = 11.84TARR103 pKa = 11.84NRR105 pKa = 11.84TARR108 pKa = 11.84NRR110 pKa = 11.84TARR113 pKa = 11.84NRR115 pKa = 11.84TAQSRR120 pKa = 11.84TARR123 pKa = 11.84NRR125 pKa = 11.84TCRR128 pKa = 11.84NRR130 pKa = 11.84TARR133 pKa = 11.84NRR135 pKa = 11.84TARR138 pKa = 11.84NRR140 pKa = 11.84TARR143 pKa = 11.84NRR145 pKa = 11.84TAQSRR150 pKa = 11.84TARR153 pKa = 11.84NRR155 pKa = 11.84TAQSRR160 pKa = 11.84TARR163 pKa = 11.84NRR165 pKa = 11.84TARR168 pKa = 11.84DD169 pKa = 2.97RR170 pKa = 11.84TARR173 pKa = 11.84NRR175 pKa = 11.84TAQSRR180 pKa = 11.84TARR183 pKa = 11.84NRR185 pKa = 11.84TARR188 pKa = 11.84DD189 pKa = 2.91RR190 pKa = 11.84TTRR193 pKa = 11.84NRR195 pKa = 11.84TARR198 pKa = 11.84NRR200 pKa = 11.84TTRR203 pKa = 11.84NRR205 pKa = 11.84TARR208 pKa = 11.84SRR210 pKa = 11.84TARR213 pKa = 11.84SRR215 pKa = 11.84TARR218 pKa = 11.84SRR220 pKa = 11.84TARR223 pKa = 11.84SRR225 pKa = 11.84TARR228 pKa = 11.84NRR230 pKa = 11.84TARR233 pKa = 11.84NRR235 pKa = 11.84TAQSRR240 pKa = 11.84TARR243 pKa = 11.84NRR245 pKa = 11.84TVRR248 pKa = 11.84NRR250 pKa = 11.84TVRR253 pKa = 11.84NRR255 pKa = 11.84TARR258 pKa = 11.84NRR260 pKa = 11.84TARR263 pKa = 11.84NRR265 pKa = 11.84TARR268 pKa = 11.84NRR270 pKa = 11.84TTRR273 pKa = 11.84NRR275 pKa = 11.84TAQSRR280 pKa = 11.84TARR283 pKa = 11.84NRR285 pKa = 11.84TAQSRR290 pKa = 11.84TARR293 pKa = 11.84NRR295 pKa = 11.84TARR298 pKa = 11.84DD299 pKa = 2.91RR300 pKa = 11.84TTRR303 pKa = 11.84NRR305 pKa = 11.84TARR308 pKa = 11.84NRR310 pKa = 11.84TTRR313 pKa = 11.84NRR315 pKa = 11.84TARR318 pKa = 11.84SRR320 pKa = 11.84TARR323 pKa = 11.84SRR325 pKa = 11.84TSRR328 pKa = 11.84NRR330 pKa = 11.84TARR333 pKa = 11.84NRR335 pKa = 11.84TAQSRR340 pKa = 11.84TARR343 pKa = 11.84NRR345 pKa = 11.84TVRR348 pKa = 11.84NRR350 pKa = 11.84TVRR353 pKa = 11.84NRR355 pKa = 11.84TARR358 pKa = 11.84NRR360 pKa = 11.84TARR363 pKa = 11.84DD364 pKa = 2.91RR365 pKa = 11.84TTRR368 pKa = 11.84NRR370 pKa = 11.84TARR373 pKa = 11.84NRR375 pKa = 11.84TARR378 pKa = 11.84SRR380 pKa = 11.84TARR383 pKa = 11.84SRR385 pKa = 11.84TARR388 pKa = 11.84NRR390 pKa = 11.84TARR393 pKa = 11.84NRR395 pKa = 11.84TAQSRR400 pKa = 11.84TARR403 pKa = 11.84NRR405 pKa = 11.84TAQSRR410 pKa = 11.84TARR413 pKa = 11.84NRR415 pKa = 11.84TARR418 pKa = 11.84NRR420 pKa = 11.84TARR423 pKa = 11.84NRR425 pKa = 11.84TGSVSVQLLQSTINRR440 pKa = 11.84RR441 pKa = 11.84LIHH444 pKa = 6.54RR445 pKa = 11.84LINRR449 pKa = 11.84LNQSTINLVDD459 pKa = 3.26
MM1 pKa = 7.46YY2 pKa = 10.32SRR4 pKa = 11.84SPPPRR9 pKa = 11.84GQLPDD14 pKa = 3.48SYY16 pKa = 11.18LAAVWRR22 pKa = 11.84LLGGSLAALWRR33 pKa = 11.84LFGGSLAALWRR44 pKa = 11.84LCVGYY49 pKa = 8.19RR50 pKa = 11.84TARR53 pKa = 11.84NRR55 pKa = 11.84TARR58 pKa = 11.84NRR60 pKa = 11.84TARR63 pKa = 11.84NRR65 pKa = 11.84TTRR68 pKa = 11.84NRR70 pKa = 11.84TARR73 pKa = 11.84NRR75 pKa = 11.84TTRR78 pKa = 11.84NRR80 pKa = 11.84TARR83 pKa = 11.84SRR85 pKa = 11.84TARR88 pKa = 11.84SRR90 pKa = 11.84TARR93 pKa = 11.84NRR95 pKa = 11.84TARR98 pKa = 11.84SRR100 pKa = 11.84TARR103 pKa = 11.84NRR105 pKa = 11.84TARR108 pKa = 11.84NRR110 pKa = 11.84TARR113 pKa = 11.84NRR115 pKa = 11.84TAQSRR120 pKa = 11.84TARR123 pKa = 11.84NRR125 pKa = 11.84TCRR128 pKa = 11.84NRR130 pKa = 11.84TARR133 pKa = 11.84NRR135 pKa = 11.84TARR138 pKa = 11.84NRR140 pKa = 11.84TARR143 pKa = 11.84NRR145 pKa = 11.84TAQSRR150 pKa = 11.84TARR153 pKa = 11.84NRR155 pKa = 11.84TAQSRR160 pKa = 11.84TARR163 pKa = 11.84NRR165 pKa = 11.84TARR168 pKa = 11.84DD169 pKa = 2.97RR170 pKa = 11.84TARR173 pKa = 11.84NRR175 pKa = 11.84TAQSRR180 pKa = 11.84TARR183 pKa = 11.84NRR185 pKa = 11.84TARR188 pKa = 11.84DD189 pKa = 2.91RR190 pKa = 11.84TTRR193 pKa = 11.84NRR195 pKa = 11.84TARR198 pKa = 11.84NRR200 pKa = 11.84TTRR203 pKa = 11.84NRR205 pKa = 11.84TARR208 pKa = 11.84SRR210 pKa = 11.84TARR213 pKa = 11.84SRR215 pKa = 11.84TARR218 pKa = 11.84SRR220 pKa = 11.84TARR223 pKa = 11.84SRR225 pKa = 11.84TARR228 pKa = 11.84NRR230 pKa = 11.84TARR233 pKa = 11.84NRR235 pKa = 11.84TAQSRR240 pKa = 11.84TARR243 pKa = 11.84NRR245 pKa = 11.84TVRR248 pKa = 11.84NRR250 pKa = 11.84TVRR253 pKa = 11.84NRR255 pKa = 11.84TARR258 pKa = 11.84NRR260 pKa = 11.84TARR263 pKa = 11.84NRR265 pKa = 11.84TARR268 pKa = 11.84NRR270 pKa = 11.84TTRR273 pKa = 11.84NRR275 pKa = 11.84TAQSRR280 pKa = 11.84TARR283 pKa = 11.84NRR285 pKa = 11.84TAQSRR290 pKa = 11.84TARR293 pKa = 11.84NRR295 pKa = 11.84TARR298 pKa = 11.84DD299 pKa = 2.91RR300 pKa = 11.84TTRR303 pKa = 11.84NRR305 pKa = 11.84TARR308 pKa = 11.84NRR310 pKa = 11.84TTRR313 pKa = 11.84NRR315 pKa = 11.84TARR318 pKa = 11.84SRR320 pKa = 11.84TARR323 pKa = 11.84SRR325 pKa = 11.84TSRR328 pKa = 11.84NRR330 pKa = 11.84TARR333 pKa = 11.84NRR335 pKa = 11.84TAQSRR340 pKa = 11.84TARR343 pKa = 11.84NRR345 pKa = 11.84TVRR348 pKa = 11.84NRR350 pKa = 11.84TVRR353 pKa = 11.84NRR355 pKa = 11.84TARR358 pKa = 11.84NRR360 pKa = 11.84TARR363 pKa = 11.84DD364 pKa = 2.91RR365 pKa = 11.84TTRR368 pKa = 11.84NRR370 pKa = 11.84TARR373 pKa = 11.84NRR375 pKa = 11.84TARR378 pKa = 11.84SRR380 pKa = 11.84TARR383 pKa = 11.84SRR385 pKa = 11.84TARR388 pKa = 11.84NRR390 pKa = 11.84TARR393 pKa = 11.84NRR395 pKa = 11.84TAQSRR400 pKa = 11.84TARR403 pKa = 11.84NRR405 pKa = 11.84TAQSRR410 pKa = 11.84TARR413 pKa = 11.84NRR415 pKa = 11.84TARR418 pKa = 11.84NRR420 pKa = 11.84TARR423 pKa = 11.84NRR425 pKa = 11.84TGSVSVQLLQSTINRR440 pKa = 11.84RR441 pKa = 11.84LIHH444 pKa = 6.54RR445 pKa = 11.84LINRR449 pKa = 11.84LNQSTINLVDD459 pKa = 3.26
Molecular weight: 53.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10578374 |
20 |
9371 |
302.0 |
34.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.83 ± 0.016 | 1.936 ± 0.012 |
5.34 ± 0.014 | 6.787 ± 0.022 |
4.307 ± 0.012 | 5.428 ± 0.017 |
2.344 ± 0.008 | 5.563 ± 0.011 |
6.323 ± 0.018 | 8.517 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.543 ± 0.007 | 4.648 ± 0.012 |
5.065 ± 0.017 | 4.12 ± 0.014 |
5.905 ± 0.019 | 7.96 ± 0.02 |
5.925 ± 0.016 | 6.356 ± 0.012 |
1.173 ± 0.005 | 2.87 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
