
Faeces associated gemycircularvirus 21
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus lamas1; Lama associated gemycircularvirus 1
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
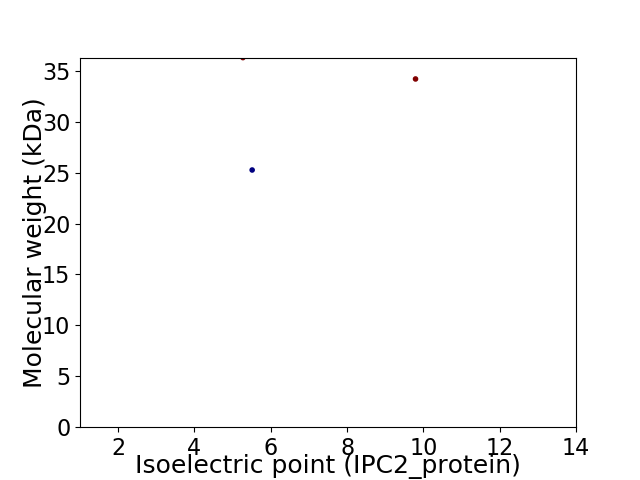
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A161IBB7|A0A161IBB7_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 21 OX=1843741 PE=3 SV=1
MM1 pKa = 7.34TFAINAKK8 pKa = 9.8YY9 pKa = 10.82VLLTYY14 pKa = 9.11AQCGDD19 pKa = 3.65LDD21 pKa = 4.01GFAVMDD27 pKa = 5.51RR28 pKa = 11.84ISEE31 pKa = 4.19LGGEE35 pKa = 4.48CIVARR40 pKa = 11.84EE41 pKa = 3.98THH43 pKa = 6.64ADD45 pKa = 3.46GGTHH49 pKa = 6.5LHH51 pKa = 5.76VFCDD55 pKa = 4.88FGRR58 pKa = 11.84KK59 pKa = 8.16FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 7.62TDD66 pKa = 2.99VFDD69 pKa = 3.6VLGYY73 pKa = 10.25HH74 pKa = 6.77PNIEE78 pKa = 4.21PSRR81 pKa = 11.84GTPEE85 pKa = 3.59KK86 pKa = 10.76GYY88 pKa = 10.89DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.31VICGGLARR104 pKa = 11.84PTTSRR109 pKa = 11.84TGDD112 pKa = 2.98SSANSKK118 pKa = 6.86WTEE121 pKa = 3.6ITSATNRR128 pKa = 11.84DD129 pKa = 3.88EE130 pKa = 4.4FWEE133 pKa = 4.4LVHH136 pKa = 8.01LLDD139 pKa = 5.09PKK141 pKa = 10.8SAACSFGQLQKK152 pKa = 10.46YY153 pKa = 9.47CDD155 pKa = 3.4WKK157 pKa = 10.91FAVAPPTYY165 pKa = 9.42TSPTGVEE172 pKa = 4.16FDD174 pKa = 4.78DD175 pKa = 5.19GSIDD179 pKa = 5.11GRR181 pKa = 11.84LDD183 pKa = 2.97WLQQSGVGSGQPLLGRR199 pKa = 11.84CMSLCLYY206 pKa = 10.73GEE208 pKa = 4.21SRR210 pKa = 11.84TGKK213 pKa = 7.35TLWARR218 pKa = 11.84SLGAHH223 pKa = 7.21IYY225 pKa = 10.48CVGLVSGDD233 pKa = 3.48EE234 pKa = 4.17CMKK237 pKa = 11.0ADD239 pKa = 4.21DD240 pKa = 4.31ADD242 pKa = 3.78YY243 pKa = 11.27AIFDD247 pKa = 5.27DD248 pKa = 3.68IRR250 pKa = 11.84GGIKK254 pKa = 9.99FFPSFKK260 pKa = 10.33EE261 pKa = 3.83WLGCQAWVTVKK272 pKa = 10.5CLYY275 pKa = 10.42RR276 pKa = 11.84EE277 pKa = 4.05PKK279 pKa = 9.36LIKK282 pKa = 9.2WGKK285 pKa = 7.39PSIWISNTDD294 pKa = 3.03PRR296 pKa = 11.84DD297 pKa = 3.62NMEE300 pKa = 4.08TSDD303 pKa = 3.87VHH305 pKa = 6.6WMNKK309 pKa = 8.26NCIFVAVDD317 pKa = 3.45SPIFRR322 pKa = 11.84ANTEE326 pKa = 3.84
MM1 pKa = 7.34TFAINAKK8 pKa = 9.8YY9 pKa = 10.82VLLTYY14 pKa = 9.11AQCGDD19 pKa = 3.65LDD21 pKa = 4.01GFAVMDD27 pKa = 5.51RR28 pKa = 11.84ISEE31 pKa = 4.19LGGEE35 pKa = 4.48CIVARR40 pKa = 11.84EE41 pKa = 3.98THH43 pKa = 6.64ADD45 pKa = 3.46GGTHH49 pKa = 6.5LHH51 pKa = 5.76VFCDD55 pKa = 4.88FGRR58 pKa = 11.84KK59 pKa = 8.16FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 7.62TDD66 pKa = 2.99VFDD69 pKa = 3.6VLGYY73 pKa = 10.25HH74 pKa = 6.77PNIEE78 pKa = 4.21PSRR81 pKa = 11.84GTPEE85 pKa = 3.59KK86 pKa = 10.76GYY88 pKa = 10.89DD89 pKa = 3.47YY90 pKa = 10.69AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.31VICGGLARR104 pKa = 11.84PTTSRR109 pKa = 11.84TGDD112 pKa = 2.98SSANSKK118 pKa = 6.86WTEE121 pKa = 3.6ITSATNRR128 pKa = 11.84DD129 pKa = 3.88EE130 pKa = 4.4FWEE133 pKa = 4.4LVHH136 pKa = 8.01LLDD139 pKa = 5.09PKK141 pKa = 10.8SAACSFGQLQKK152 pKa = 10.46YY153 pKa = 9.47CDD155 pKa = 3.4WKK157 pKa = 10.91FAVAPPTYY165 pKa = 9.42TSPTGVEE172 pKa = 4.16FDD174 pKa = 4.78DD175 pKa = 5.19GSIDD179 pKa = 5.11GRR181 pKa = 11.84LDD183 pKa = 2.97WLQQSGVGSGQPLLGRR199 pKa = 11.84CMSLCLYY206 pKa = 10.73GEE208 pKa = 4.21SRR210 pKa = 11.84TGKK213 pKa = 7.35TLWARR218 pKa = 11.84SLGAHH223 pKa = 7.21IYY225 pKa = 10.48CVGLVSGDD233 pKa = 3.48EE234 pKa = 4.17CMKK237 pKa = 11.0ADD239 pKa = 4.21DD240 pKa = 4.31ADD242 pKa = 3.78YY243 pKa = 11.27AIFDD247 pKa = 5.27DD248 pKa = 3.68IRR250 pKa = 11.84GGIKK254 pKa = 9.99FFPSFKK260 pKa = 10.33EE261 pKa = 3.83WLGCQAWVTVKK272 pKa = 10.5CLYY275 pKa = 10.42RR276 pKa = 11.84EE277 pKa = 4.05PKK279 pKa = 9.36LIKK282 pKa = 9.2WGKK285 pKa = 7.39PSIWISNTDD294 pKa = 3.03PRR296 pKa = 11.84DD297 pKa = 3.62NMEE300 pKa = 4.08TSDD303 pKa = 3.87VHH305 pKa = 6.6WMNKK309 pKa = 8.26NCIFVAVDD317 pKa = 3.45SPIFRR322 pKa = 11.84ANTEE326 pKa = 3.84
Molecular weight: 36.29 kDa
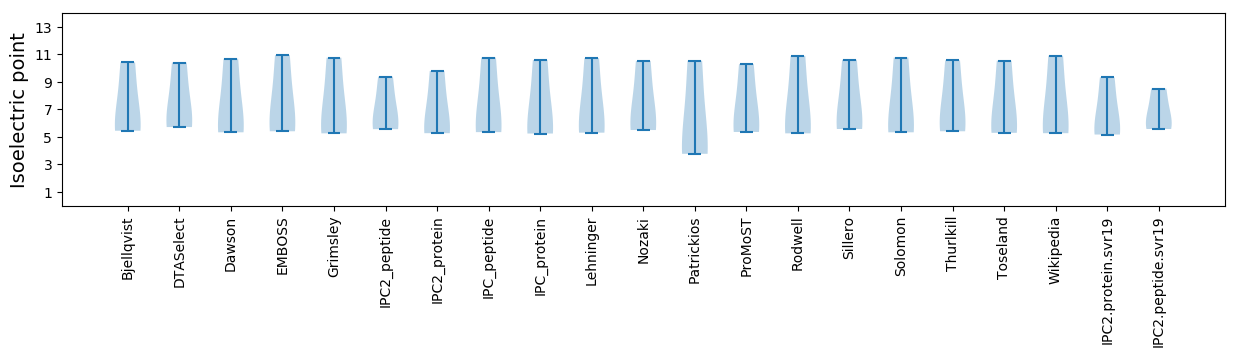
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161IBB7|A0A161IBB7_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 21 OX=1843741 PE=3 SV=1
MM1 pKa = 7.75AYY3 pKa = 10.21ARR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84FPRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AFKK16 pKa = 10.3RR17 pKa = 11.84RR18 pKa = 11.84SAPRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 7.59VAKK30 pKa = 10.17RR31 pKa = 11.84PSRR34 pKa = 11.84TRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84AMPSRR43 pKa = 11.84RR44 pKa = 11.84SILNLTTTKK53 pKa = 10.42KK54 pKa = 9.85RR55 pKa = 11.84DD56 pKa = 3.28NMAPAFYY63 pKa = 10.19RR64 pKa = 11.84AGDD67 pKa = 3.89GVTATPVNNGIALLATMPAYY87 pKa = 7.19TTAHH91 pKa = 5.96CVLWCATEE99 pKa = 4.17RR100 pKa = 11.84EE101 pKa = 4.42YY102 pKa = 11.52ASGSNMSDD110 pKa = 2.72HH111 pKa = 6.77GRR113 pKa = 11.84RR114 pKa = 11.84SHH116 pKa = 5.45TPFYY120 pKa = 10.51KK121 pKa = 10.64GLSEE125 pKa = 5.04RR126 pKa = 11.84IRR128 pKa = 11.84MKK130 pKa = 8.59TTNSSPWLWRR140 pKa = 11.84RR141 pKa = 11.84IVFTMKK147 pKa = 10.56GPLPEE152 pKa = 4.61TIRR155 pKa = 11.84TSIKK159 pKa = 10.38VGNQYY164 pKa = 10.67KK165 pKa = 10.44RR166 pKa = 11.84GITWLTDD173 pKa = 3.28GSDD176 pKa = 3.11VGNSVNGLVNTTLFAGSAGIDD197 pKa = 3.43WNDD200 pKa = 2.91VMLAKK205 pKa = 9.95TDD207 pKa = 3.8TQALTVMYY215 pKa = 10.45DD216 pKa = 2.99RR217 pKa = 11.84TIQLKK222 pKa = 10.35SGSDD226 pKa = 3.09SGTIRR231 pKa = 11.84DD232 pKa = 4.43FKK234 pKa = 10.45MCHH237 pKa = 6.06PMNKK241 pKa = 8.1TLVYY245 pKa = 10.28AYY247 pKa = 10.02DD248 pKa = 3.46EE249 pKa = 4.77SGTFDD254 pKa = 3.22VSFTRR259 pKa = 11.84SALGRR264 pKa = 11.84AGMGDD269 pKa = 3.44MYY271 pKa = 10.18IYY273 pKa = 11.32DD274 pKa = 4.12MFQPHH279 pKa = 7.26DD280 pKa = 3.85FSVDD284 pKa = 2.88GGTIEE289 pKa = 4.65FNPKK293 pKa = 8.25ATLYY297 pKa = 9.09WHH299 pKa = 6.97EE300 pKa = 4.28KK301 pKa = 9.3
MM1 pKa = 7.75AYY3 pKa = 10.21ARR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84FPRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AFKK16 pKa = 10.3RR17 pKa = 11.84RR18 pKa = 11.84SAPRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 7.59VAKK30 pKa = 10.17RR31 pKa = 11.84PSRR34 pKa = 11.84TRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84AMPSRR43 pKa = 11.84RR44 pKa = 11.84SILNLTTTKK53 pKa = 10.42KK54 pKa = 9.85RR55 pKa = 11.84DD56 pKa = 3.28NMAPAFYY63 pKa = 10.19RR64 pKa = 11.84AGDD67 pKa = 3.89GVTATPVNNGIALLATMPAYY87 pKa = 7.19TTAHH91 pKa = 5.96CVLWCATEE99 pKa = 4.17RR100 pKa = 11.84EE101 pKa = 4.42YY102 pKa = 11.52ASGSNMSDD110 pKa = 2.72HH111 pKa = 6.77GRR113 pKa = 11.84RR114 pKa = 11.84SHH116 pKa = 5.45TPFYY120 pKa = 10.51KK121 pKa = 10.64GLSEE125 pKa = 5.04RR126 pKa = 11.84IRR128 pKa = 11.84MKK130 pKa = 8.59TTNSSPWLWRR140 pKa = 11.84RR141 pKa = 11.84IVFTMKK147 pKa = 10.56GPLPEE152 pKa = 4.61TIRR155 pKa = 11.84TSIKK159 pKa = 10.38VGNQYY164 pKa = 10.67KK165 pKa = 10.44RR166 pKa = 11.84GITWLTDD173 pKa = 3.28GSDD176 pKa = 3.11VGNSVNGLVNTTLFAGSAGIDD197 pKa = 3.43WNDD200 pKa = 2.91VMLAKK205 pKa = 9.95TDD207 pKa = 3.8TQALTVMYY215 pKa = 10.45DD216 pKa = 2.99RR217 pKa = 11.84TIQLKK222 pKa = 10.35SGSDD226 pKa = 3.09SGTIRR231 pKa = 11.84DD232 pKa = 4.43FKK234 pKa = 10.45MCHH237 pKa = 6.06PMNKK241 pKa = 8.1TLVYY245 pKa = 10.28AYY247 pKa = 10.02DD248 pKa = 3.46EE249 pKa = 4.77SGTFDD254 pKa = 3.22VSFTRR259 pKa = 11.84SALGRR264 pKa = 11.84AGMGDD269 pKa = 3.44MYY271 pKa = 10.18IYY273 pKa = 11.32DD274 pKa = 4.12MFQPHH279 pKa = 7.26DD280 pKa = 3.85FSVDD284 pKa = 2.88GGTIEE289 pKa = 4.65FNPKK293 pKa = 8.25ATLYY297 pKa = 9.09WHH299 pKa = 6.97EE300 pKa = 4.28KK301 pKa = 9.3
Molecular weight: 34.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
860 |
233 |
326 |
286.7 |
31.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.093 ± 0.403 | 2.674 ± 0.793 |
7.442 ± 0.818 | 3.605 ± 0.604 |
4.419 ± 0.242 | 9.186 ± 0.924 |
2.093 ± 0.045 | 4.419 ± 0.367 |
5.0 ± 0.281 | 7.093 ± 0.786 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.558 ± 0.969 | 3.023 ± 0.629 |
4.884 ± 0.404 | 1.977 ± 0.307 |
7.558 ± 1.701 | 7.674 ± 0.287 |
8.023 ± 0.929 | 5.349 ± 0.333 |
2.326 ± 0.345 | 3.605 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
