
Avian leukosis virus RSA (RSV-SRA) (Rous sarcoma virus (strain Schmidt-Ruppin A))
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Alpharetrovirus; Rous sarcoma virus; Rous sarcoma virus (strain Schmidt-Ruppin)
Average proteome isoelectric point is 7.88
Get precalculated fractions of proteins
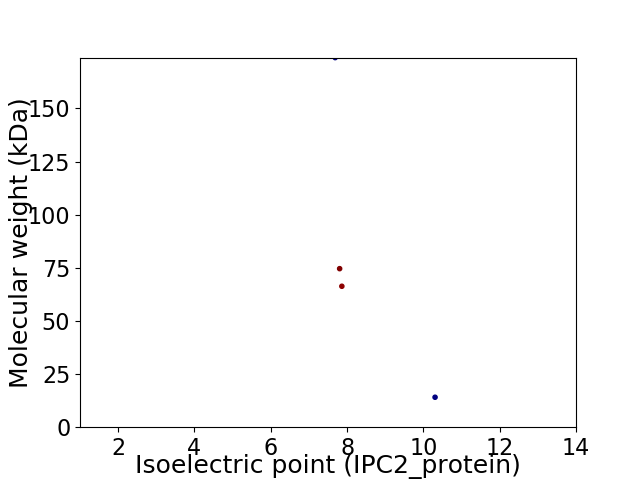
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
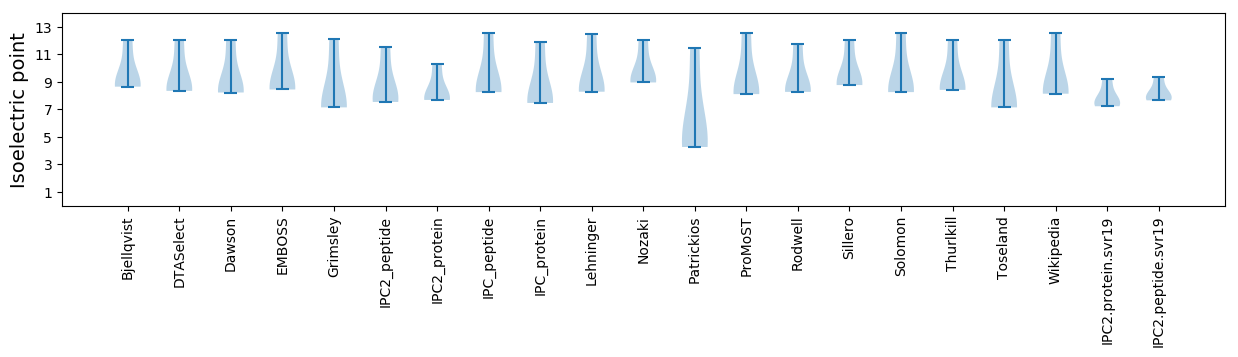
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q04221|VP14_RSVSA Putative trans-acting factor OS=Avian leukosis virus RSA OX=269446 GN=trans-acting factor PE=4 SV=1
MM1 pKa = 7.24EE2 pKa = 5.23AVIKK6 pKa = 10.33VISSACKK13 pKa = 8.59TYY15 pKa = 10.41CGKK18 pKa = 10.33ISPSKK23 pKa = 10.7KK24 pKa = 9.94EE25 pKa = 3.54IGAMLSLLQKK35 pKa = 10.32EE36 pKa = 4.76GLLMSPSDD44 pKa = 4.13LYY46 pKa = 11.72SPGSWDD52 pKa = 4.68PITAALSQRR61 pKa = 11.84AMVLGKK67 pKa = 10.37SGEE70 pKa = 4.13LKK72 pKa = 9.42TWGLVLGALKK82 pKa = 10.28AARR85 pKa = 11.84EE86 pKa = 4.03EE87 pKa = 4.39QVTSEE92 pKa = 4.35QAKK95 pKa = 9.35FWLGLGGGRR104 pKa = 11.84VSPPGPEE111 pKa = 4.63CIEE114 pKa = 4.09KK115 pKa = 9.9PATEE119 pKa = 3.75RR120 pKa = 11.84RR121 pKa = 11.84IDD123 pKa = 3.28KK124 pKa = 10.81GEE126 pKa = 3.96EE127 pKa = 3.89VGEE130 pKa = 4.28TTAQRR135 pKa = 11.84DD136 pKa = 3.37AKK138 pKa = 9.74MAPEE142 pKa = 4.28KK143 pKa = 10.19MATPKK148 pKa = 10.17TVGTSCYY155 pKa = 10.07QCGTATGCNCATASAPPPPYY175 pKa = 9.97VGSGLYY181 pKa = 9.82PSLAGVGEE189 pKa = 4.04QQGQGGDD196 pKa = 3.96TPWGAEE202 pKa = 3.88QPRR205 pKa = 11.84AEE207 pKa = 4.76PGHH210 pKa = 6.9AGLAPGPALTDD221 pKa = 3.15WARR224 pKa = 11.84IRR226 pKa = 11.84EE227 pKa = 4.27EE228 pKa = 4.12LASTGPPVVAMPVVIKK244 pKa = 9.98TEE246 pKa = 4.41GPAWTPLEE254 pKa = 4.33PKK256 pKa = 10.46LITRR260 pKa = 11.84LADD263 pKa = 3.44TVRR266 pKa = 11.84TKK268 pKa = 11.01GLRR271 pKa = 11.84SPITMAEE278 pKa = 4.01VEE280 pKa = 4.65ALMSSPLLPHH290 pKa = 7.26DD291 pKa = 3.78VTNLMRR297 pKa = 11.84VILGPAPYY305 pKa = 9.99ALWMDD310 pKa = 3.45AWGVQLQTVIAAATRR325 pKa = 11.84DD326 pKa = 3.68PRR328 pKa = 11.84HH329 pKa = 6.07PANGQGRR336 pKa = 11.84GEE338 pKa = 4.12RR339 pKa = 11.84TNLDD343 pKa = 3.72RR344 pKa = 11.84LKK346 pKa = 11.02GLADD350 pKa = 3.43GMVGNPQGQAALLRR364 pKa = 11.84PGEE367 pKa = 4.17LVAITASALQAFRR380 pKa = 11.84EE381 pKa = 4.5VARR384 pKa = 11.84LAEE387 pKa = 4.33PAGPWADD394 pKa = 3.01ITQGPSEE401 pKa = 4.53SFVDD405 pKa = 4.25FANRR409 pKa = 11.84LIKK412 pKa = 10.44AVEE415 pKa = 4.43GSDD418 pKa = 3.86LPPSARR424 pKa = 11.84APVIIDD430 pKa = 3.54CFRR433 pKa = 11.84QKK435 pKa = 10.34SQPDD439 pKa = 3.21IQQLIRR445 pKa = 11.84AAPSTLTTPGEE456 pKa = 4.01IIKK459 pKa = 10.7YY460 pKa = 9.86VLDD463 pKa = 3.78RR464 pKa = 11.84QKK466 pKa = 10.46IAPLTDD472 pKa = 3.04QGIAAAMSSAIQPLVMAVVNRR493 pKa = 11.84EE494 pKa = 3.55RR495 pKa = 11.84DD496 pKa = 3.61GQTGSGGRR504 pKa = 11.84ARR506 pKa = 11.84GLCYY510 pKa = 9.87TCGSPGHH517 pKa = 5.93YY518 pKa = 9.31QAQCPKK524 pKa = 9.91KK525 pKa = 9.55RR526 pKa = 11.84KK527 pKa = 9.3SGNSRR532 pKa = 11.84EE533 pKa = 3.99RR534 pKa = 11.84CQLCDD539 pKa = 4.32GMGHH543 pKa = 6.08NAKK546 pKa = 9.54QCRR549 pKa = 11.84RR550 pKa = 11.84RR551 pKa = 11.84DD552 pKa = 3.51GNQGQRR558 pKa = 11.84PGKK561 pKa = 9.73GLSSGSWPVSEE572 pKa = 4.54QPAVSLAMTMEE583 pKa = 4.56HH584 pKa = 7.35KK585 pKa = 10.49DD586 pKa = 3.43RR587 pKa = 11.84PLVRR591 pKa = 11.84VILTNTGSHH600 pKa = 6.33PVKK603 pKa = 10.3QRR605 pKa = 11.84SVYY608 pKa = 8.6ITALLDD614 pKa = 3.34SGADD618 pKa = 2.97ITIISEE624 pKa = 4.22EE625 pKa = 4.3DD626 pKa = 3.45WPTDD630 pKa = 3.33WPVMEE635 pKa = 4.92AANPQIHH642 pKa = 6.88GIGGGIPMRR651 pKa = 11.84KK652 pKa = 8.45SRR654 pKa = 11.84DD655 pKa = 3.55MIEE658 pKa = 3.91VGVINRR664 pKa = 11.84DD665 pKa = 3.23GSLEE669 pKa = 4.04RR670 pKa = 11.84PLLLFPAVAMVRR682 pKa = 11.84GSILGRR688 pKa = 11.84DD689 pKa = 4.06CLQGLGLRR697 pKa = 11.84LTNLIGRR704 pKa = 11.84ATVLTVALHH713 pKa = 6.18LAIPLKK719 pKa = 9.84WKK721 pKa = 9.73PDD723 pKa = 3.61HH724 pKa = 6.02TPVWIDD730 pKa = 2.94QWPLPEE736 pKa = 4.87GKK738 pKa = 10.01LVALTQLVEE747 pKa = 4.57KK748 pKa = 9.81EE749 pKa = 4.09LQLGHH754 pKa = 6.94IEE756 pKa = 4.41PSLSCWNTPVFVIRR770 pKa = 11.84KK771 pKa = 8.86ASGSYY776 pKa = 10.32RR777 pKa = 11.84LLHH780 pKa = 7.0DD781 pKa = 4.61LRR783 pKa = 11.84AVNAKK788 pKa = 9.53LVPFGAVQQGAPVLSALPRR807 pKa = 11.84GWPLMVLDD815 pKa = 5.46LKK817 pKa = 11.31DD818 pKa = 4.03CFFSIPLAEE827 pKa = 4.19QDD829 pKa = 3.37RR830 pKa = 11.84EE831 pKa = 4.28AFAFTLPSVNNQAPARR847 pKa = 11.84RR848 pKa = 11.84FQWKK852 pKa = 9.35VLPQGMTCSPTICQLVVGQVLEE874 pKa = 4.13PLRR877 pKa = 11.84LKK879 pKa = 10.8HH880 pKa = 6.03PSLRR884 pKa = 11.84MLHH887 pKa = 6.24YY888 pKa = 10.21MDD890 pKa = 6.17DD891 pKa = 4.09LLLAASSHH899 pKa = 7.13DD900 pKa = 3.55GLEE903 pKa = 4.16AAGEE907 pKa = 4.24EE908 pKa = 4.56VISTLEE914 pKa = 3.75RR915 pKa = 11.84AGFTISPDD923 pKa = 3.32KK924 pKa = 10.49IQRR927 pKa = 11.84EE928 pKa = 4.07PGVQYY933 pKa = 10.87LGYY936 pKa = 10.0KK937 pKa = 10.18LGSTYY942 pKa = 10.3VAPVGLVAEE951 pKa = 4.42PRR953 pKa = 11.84IATLWDD959 pKa = 3.32VQKK962 pKa = 11.12LVGSLQWLRR971 pKa = 11.84PALGIPPRR979 pKa = 11.84LMGPFYY985 pKa = 10.43EE986 pKa = 4.39QLRR989 pKa = 11.84GSDD992 pKa = 3.66PNEE995 pKa = 3.45ARR997 pKa = 11.84EE998 pKa = 4.15WNLDD1002 pKa = 2.98MKK1004 pKa = 9.5MAWRR1008 pKa = 11.84EE1009 pKa = 3.71IVQLSTTAALEE1020 pKa = 4.09RR1021 pKa = 11.84WDD1023 pKa = 3.82PALPLEE1029 pKa = 4.65GAVARR1034 pKa = 11.84CEE1036 pKa = 3.77QGAIGVLGQGLSTHH1050 pKa = 6.51PRR1052 pKa = 11.84PCLWLFSTQPTKK1064 pKa = 11.17AFTAWLEE1071 pKa = 4.03VLTLLITKK1079 pKa = 9.91LRR1081 pKa = 11.84ASAVRR1086 pKa = 11.84TFGKK1090 pKa = 10.42EE1091 pKa = 3.51VDD1093 pKa = 3.67ILLLPACFRR1102 pKa = 11.84EE1103 pKa = 4.37DD1104 pKa = 3.38LPLPEE1109 pKa = 5.79GILLALRR1116 pKa = 11.84GFAGKK1121 pKa = 9.52IRR1123 pKa = 11.84SSDD1126 pKa = 3.42TPSIFDD1132 pKa = 3.27IARR1135 pKa = 11.84PLHH1138 pKa = 5.93VSLKK1142 pKa = 10.28VRR1144 pKa = 11.84VTDD1147 pKa = 3.89HH1148 pKa = 6.83PVPGPTAFTDD1158 pKa = 3.96ASSSTHH1164 pKa = 5.96KK1165 pKa = 10.92GVVVWRR1171 pKa = 11.84EE1172 pKa = 3.75GPRR1175 pKa = 11.84WEE1177 pKa = 4.22IKK1179 pKa = 10.37EE1180 pKa = 3.98IADD1183 pKa = 4.09LGASVQQLEE1192 pKa = 4.18ARR1194 pKa = 11.84AVAMALLLWPTTPTNVVTDD1213 pKa = 3.73SAFVAKK1219 pKa = 9.61MLLKK1223 pKa = 9.82MGQEE1227 pKa = 4.63GVPSTAAAFILEE1239 pKa = 4.5DD1240 pKa = 3.69ALSQRR1245 pKa = 11.84SAMAAVLHH1253 pKa = 5.73VRR1255 pKa = 11.84SHH1257 pKa = 7.02SEE1259 pKa = 3.69VPGFFTEE1266 pKa = 4.54GNDD1269 pKa = 3.71VADD1272 pKa = 3.87SQATFQAYY1280 pKa = 8.62PLRR1283 pKa = 11.84EE1284 pKa = 4.2AKK1286 pKa = 10.14DD1287 pKa = 3.34LHH1289 pKa = 5.98TALHH1293 pKa = 6.29IGPRR1297 pKa = 11.84ALSKK1301 pKa = 10.84ACNISMQQARR1311 pKa = 11.84EE1312 pKa = 4.09VVQTCPHH1319 pKa = 6.59CNSAPALEE1327 pKa = 4.92AGVNPRR1333 pKa = 11.84GLGPLQIWQTDD1344 pKa = 3.79FTLEE1348 pKa = 3.58PRR1350 pKa = 11.84MAPRR1354 pKa = 11.84SWLAVTVDD1362 pKa = 3.75TASSAIVVTQHH1373 pKa = 5.89GRR1375 pKa = 11.84VTSVAAQHH1383 pKa = 5.86HH1384 pKa = 6.5WATAIAVLGRR1394 pKa = 11.84PKK1396 pKa = 10.5AIKK1399 pKa = 9.02TDD1401 pKa = 3.34NGSCFTSKK1409 pKa = 8.57STRR1412 pKa = 11.84EE1413 pKa = 3.52WLARR1417 pKa = 11.84WGIAHH1422 pKa = 5.78TTGIPGNSQGQAMVEE1437 pKa = 3.89RR1438 pKa = 11.84ANRR1441 pKa = 11.84LLKK1444 pKa = 10.84DD1445 pKa = 3.88KK1446 pKa = 10.85IRR1448 pKa = 11.84VLAEE1452 pKa = 3.57GDD1454 pKa = 3.48GFMKK1458 pKa = 10.6RR1459 pKa = 11.84IPTSKK1464 pKa = 10.19QGEE1467 pKa = 4.36LLAKK1471 pKa = 10.45AMYY1474 pKa = 10.28ALNHH1478 pKa = 6.25FEE1480 pKa = 4.52RR1481 pKa = 11.84GEE1483 pKa = 4.08NTKK1486 pKa = 9.6TPIQKK1491 pKa = 9.21HH1492 pKa = 4.25WRR1494 pKa = 11.84PTVLTEE1500 pKa = 4.11GPPVKK1505 pKa = 10.12IRR1507 pKa = 11.84IEE1509 pKa = 3.85TGEE1512 pKa = 4.09WEE1514 pKa = 4.34KK1515 pKa = 11.25GWNVLVWGRR1524 pKa = 11.84GYY1526 pKa = 11.12AAVKK1530 pKa = 10.59NRR1532 pKa = 11.84DD1533 pKa = 3.13TDD1535 pKa = 3.61KK1536 pKa = 11.36VIWVPSRR1543 pKa = 11.84KK1544 pKa = 9.39VKK1546 pKa = 10.52PDD1548 pKa = 3.13VTQKK1552 pKa = 11.49DD1553 pKa = 3.91EE1554 pKa = 4.15VTKK1557 pKa = 10.61KK1558 pKa = 10.9DD1559 pKa = 3.32EE1560 pKa = 4.5ASPLFAGISDD1570 pKa = 4.51WIPWEE1575 pKa = 4.39DD1576 pKa = 3.79EE1577 pKa = 4.05QEE1579 pKa = 4.24GLQGEE1584 pKa = 4.55TASNKK1589 pKa = 7.35QEE1591 pKa = 4.17RR1592 pKa = 11.84PGEE1595 pKa = 4.05DD1596 pKa = 2.5TLAANEE1602 pKa = 4.27SS1603 pKa = 3.7
MM1 pKa = 7.24EE2 pKa = 5.23AVIKK6 pKa = 10.33VISSACKK13 pKa = 8.59TYY15 pKa = 10.41CGKK18 pKa = 10.33ISPSKK23 pKa = 10.7KK24 pKa = 9.94EE25 pKa = 3.54IGAMLSLLQKK35 pKa = 10.32EE36 pKa = 4.76GLLMSPSDD44 pKa = 4.13LYY46 pKa = 11.72SPGSWDD52 pKa = 4.68PITAALSQRR61 pKa = 11.84AMVLGKK67 pKa = 10.37SGEE70 pKa = 4.13LKK72 pKa = 9.42TWGLVLGALKK82 pKa = 10.28AARR85 pKa = 11.84EE86 pKa = 4.03EE87 pKa = 4.39QVTSEE92 pKa = 4.35QAKK95 pKa = 9.35FWLGLGGGRR104 pKa = 11.84VSPPGPEE111 pKa = 4.63CIEE114 pKa = 4.09KK115 pKa = 9.9PATEE119 pKa = 3.75RR120 pKa = 11.84RR121 pKa = 11.84IDD123 pKa = 3.28KK124 pKa = 10.81GEE126 pKa = 3.96EE127 pKa = 3.89VGEE130 pKa = 4.28TTAQRR135 pKa = 11.84DD136 pKa = 3.37AKK138 pKa = 9.74MAPEE142 pKa = 4.28KK143 pKa = 10.19MATPKK148 pKa = 10.17TVGTSCYY155 pKa = 10.07QCGTATGCNCATASAPPPPYY175 pKa = 9.97VGSGLYY181 pKa = 9.82PSLAGVGEE189 pKa = 4.04QQGQGGDD196 pKa = 3.96TPWGAEE202 pKa = 3.88QPRR205 pKa = 11.84AEE207 pKa = 4.76PGHH210 pKa = 6.9AGLAPGPALTDD221 pKa = 3.15WARR224 pKa = 11.84IRR226 pKa = 11.84EE227 pKa = 4.27EE228 pKa = 4.12LASTGPPVVAMPVVIKK244 pKa = 9.98TEE246 pKa = 4.41GPAWTPLEE254 pKa = 4.33PKK256 pKa = 10.46LITRR260 pKa = 11.84LADD263 pKa = 3.44TVRR266 pKa = 11.84TKK268 pKa = 11.01GLRR271 pKa = 11.84SPITMAEE278 pKa = 4.01VEE280 pKa = 4.65ALMSSPLLPHH290 pKa = 7.26DD291 pKa = 3.78VTNLMRR297 pKa = 11.84VILGPAPYY305 pKa = 9.99ALWMDD310 pKa = 3.45AWGVQLQTVIAAATRR325 pKa = 11.84DD326 pKa = 3.68PRR328 pKa = 11.84HH329 pKa = 6.07PANGQGRR336 pKa = 11.84GEE338 pKa = 4.12RR339 pKa = 11.84TNLDD343 pKa = 3.72RR344 pKa = 11.84LKK346 pKa = 11.02GLADD350 pKa = 3.43GMVGNPQGQAALLRR364 pKa = 11.84PGEE367 pKa = 4.17LVAITASALQAFRR380 pKa = 11.84EE381 pKa = 4.5VARR384 pKa = 11.84LAEE387 pKa = 4.33PAGPWADD394 pKa = 3.01ITQGPSEE401 pKa = 4.53SFVDD405 pKa = 4.25FANRR409 pKa = 11.84LIKK412 pKa = 10.44AVEE415 pKa = 4.43GSDD418 pKa = 3.86LPPSARR424 pKa = 11.84APVIIDD430 pKa = 3.54CFRR433 pKa = 11.84QKK435 pKa = 10.34SQPDD439 pKa = 3.21IQQLIRR445 pKa = 11.84AAPSTLTTPGEE456 pKa = 4.01IIKK459 pKa = 10.7YY460 pKa = 9.86VLDD463 pKa = 3.78RR464 pKa = 11.84QKK466 pKa = 10.46IAPLTDD472 pKa = 3.04QGIAAAMSSAIQPLVMAVVNRR493 pKa = 11.84EE494 pKa = 3.55RR495 pKa = 11.84DD496 pKa = 3.61GQTGSGGRR504 pKa = 11.84ARR506 pKa = 11.84GLCYY510 pKa = 9.87TCGSPGHH517 pKa = 5.93YY518 pKa = 9.31QAQCPKK524 pKa = 9.91KK525 pKa = 9.55RR526 pKa = 11.84KK527 pKa = 9.3SGNSRR532 pKa = 11.84EE533 pKa = 3.99RR534 pKa = 11.84CQLCDD539 pKa = 4.32GMGHH543 pKa = 6.08NAKK546 pKa = 9.54QCRR549 pKa = 11.84RR550 pKa = 11.84RR551 pKa = 11.84DD552 pKa = 3.51GNQGQRR558 pKa = 11.84PGKK561 pKa = 9.73GLSSGSWPVSEE572 pKa = 4.54QPAVSLAMTMEE583 pKa = 4.56HH584 pKa = 7.35KK585 pKa = 10.49DD586 pKa = 3.43RR587 pKa = 11.84PLVRR591 pKa = 11.84VILTNTGSHH600 pKa = 6.33PVKK603 pKa = 10.3QRR605 pKa = 11.84SVYY608 pKa = 8.6ITALLDD614 pKa = 3.34SGADD618 pKa = 2.97ITIISEE624 pKa = 4.22EE625 pKa = 4.3DD626 pKa = 3.45WPTDD630 pKa = 3.33WPVMEE635 pKa = 4.92AANPQIHH642 pKa = 6.88GIGGGIPMRR651 pKa = 11.84KK652 pKa = 8.45SRR654 pKa = 11.84DD655 pKa = 3.55MIEE658 pKa = 3.91VGVINRR664 pKa = 11.84DD665 pKa = 3.23GSLEE669 pKa = 4.04RR670 pKa = 11.84PLLLFPAVAMVRR682 pKa = 11.84GSILGRR688 pKa = 11.84DD689 pKa = 4.06CLQGLGLRR697 pKa = 11.84LTNLIGRR704 pKa = 11.84ATVLTVALHH713 pKa = 6.18LAIPLKK719 pKa = 9.84WKK721 pKa = 9.73PDD723 pKa = 3.61HH724 pKa = 6.02TPVWIDD730 pKa = 2.94QWPLPEE736 pKa = 4.87GKK738 pKa = 10.01LVALTQLVEE747 pKa = 4.57KK748 pKa = 9.81EE749 pKa = 4.09LQLGHH754 pKa = 6.94IEE756 pKa = 4.41PSLSCWNTPVFVIRR770 pKa = 11.84KK771 pKa = 8.86ASGSYY776 pKa = 10.32RR777 pKa = 11.84LLHH780 pKa = 7.0DD781 pKa = 4.61LRR783 pKa = 11.84AVNAKK788 pKa = 9.53LVPFGAVQQGAPVLSALPRR807 pKa = 11.84GWPLMVLDD815 pKa = 5.46LKK817 pKa = 11.31DD818 pKa = 4.03CFFSIPLAEE827 pKa = 4.19QDD829 pKa = 3.37RR830 pKa = 11.84EE831 pKa = 4.28AFAFTLPSVNNQAPARR847 pKa = 11.84RR848 pKa = 11.84FQWKK852 pKa = 9.35VLPQGMTCSPTICQLVVGQVLEE874 pKa = 4.13PLRR877 pKa = 11.84LKK879 pKa = 10.8HH880 pKa = 6.03PSLRR884 pKa = 11.84MLHH887 pKa = 6.24YY888 pKa = 10.21MDD890 pKa = 6.17DD891 pKa = 4.09LLLAASSHH899 pKa = 7.13DD900 pKa = 3.55GLEE903 pKa = 4.16AAGEE907 pKa = 4.24EE908 pKa = 4.56VISTLEE914 pKa = 3.75RR915 pKa = 11.84AGFTISPDD923 pKa = 3.32KK924 pKa = 10.49IQRR927 pKa = 11.84EE928 pKa = 4.07PGVQYY933 pKa = 10.87LGYY936 pKa = 10.0KK937 pKa = 10.18LGSTYY942 pKa = 10.3VAPVGLVAEE951 pKa = 4.42PRR953 pKa = 11.84IATLWDD959 pKa = 3.32VQKK962 pKa = 11.12LVGSLQWLRR971 pKa = 11.84PALGIPPRR979 pKa = 11.84LMGPFYY985 pKa = 10.43EE986 pKa = 4.39QLRR989 pKa = 11.84GSDD992 pKa = 3.66PNEE995 pKa = 3.45ARR997 pKa = 11.84EE998 pKa = 4.15WNLDD1002 pKa = 2.98MKK1004 pKa = 9.5MAWRR1008 pKa = 11.84EE1009 pKa = 3.71IVQLSTTAALEE1020 pKa = 4.09RR1021 pKa = 11.84WDD1023 pKa = 3.82PALPLEE1029 pKa = 4.65GAVARR1034 pKa = 11.84CEE1036 pKa = 3.77QGAIGVLGQGLSTHH1050 pKa = 6.51PRR1052 pKa = 11.84PCLWLFSTQPTKK1064 pKa = 11.17AFTAWLEE1071 pKa = 4.03VLTLLITKK1079 pKa = 9.91LRR1081 pKa = 11.84ASAVRR1086 pKa = 11.84TFGKK1090 pKa = 10.42EE1091 pKa = 3.51VDD1093 pKa = 3.67ILLLPACFRR1102 pKa = 11.84EE1103 pKa = 4.37DD1104 pKa = 3.38LPLPEE1109 pKa = 5.79GILLALRR1116 pKa = 11.84GFAGKK1121 pKa = 9.52IRR1123 pKa = 11.84SSDD1126 pKa = 3.42TPSIFDD1132 pKa = 3.27IARR1135 pKa = 11.84PLHH1138 pKa = 5.93VSLKK1142 pKa = 10.28VRR1144 pKa = 11.84VTDD1147 pKa = 3.89HH1148 pKa = 6.83PVPGPTAFTDD1158 pKa = 3.96ASSSTHH1164 pKa = 5.96KK1165 pKa = 10.92GVVVWRR1171 pKa = 11.84EE1172 pKa = 3.75GPRR1175 pKa = 11.84WEE1177 pKa = 4.22IKK1179 pKa = 10.37EE1180 pKa = 3.98IADD1183 pKa = 4.09LGASVQQLEE1192 pKa = 4.18ARR1194 pKa = 11.84AVAMALLLWPTTPTNVVTDD1213 pKa = 3.73SAFVAKK1219 pKa = 9.61MLLKK1223 pKa = 9.82MGQEE1227 pKa = 4.63GVPSTAAAFILEE1239 pKa = 4.5DD1240 pKa = 3.69ALSQRR1245 pKa = 11.84SAMAAVLHH1253 pKa = 5.73VRR1255 pKa = 11.84SHH1257 pKa = 7.02SEE1259 pKa = 3.69VPGFFTEE1266 pKa = 4.54GNDD1269 pKa = 3.71VADD1272 pKa = 3.87SQATFQAYY1280 pKa = 8.62PLRR1283 pKa = 11.84EE1284 pKa = 4.2AKK1286 pKa = 10.14DD1287 pKa = 3.34LHH1289 pKa = 5.98TALHH1293 pKa = 6.29IGPRR1297 pKa = 11.84ALSKK1301 pKa = 10.84ACNISMQQARR1311 pKa = 11.84EE1312 pKa = 4.09VVQTCPHH1319 pKa = 6.59CNSAPALEE1327 pKa = 4.92AGVNPRR1333 pKa = 11.84GLGPLQIWQTDD1344 pKa = 3.79FTLEE1348 pKa = 3.58PRR1350 pKa = 11.84MAPRR1354 pKa = 11.84SWLAVTVDD1362 pKa = 3.75TASSAIVVTQHH1373 pKa = 5.89GRR1375 pKa = 11.84VTSVAAQHH1383 pKa = 5.86HH1384 pKa = 6.5WATAIAVLGRR1394 pKa = 11.84PKK1396 pKa = 10.5AIKK1399 pKa = 9.02TDD1401 pKa = 3.34NGSCFTSKK1409 pKa = 8.57STRR1412 pKa = 11.84EE1413 pKa = 3.52WLARR1417 pKa = 11.84WGIAHH1422 pKa = 5.78TTGIPGNSQGQAMVEE1437 pKa = 3.89RR1438 pKa = 11.84ANRR1441 pKa = 11.84LLKK1444 pKa = 10.84DD1445 pKa = 3.88KK1446 pKa = 10.85IRR1448 pKa = 11.84VLAEE1452 pKa = 3.57GDD1454 pKa = 3.48GFMKK1458 pKa = 10.6RR1459 pKa = 11.84IPTSKK1464 pKa = 10.19QGEE1467 pKa = 4.36LLAKK1471 pKa = 10.45AMYY1474 pKa = 10.28ALNHH1478 pKa = 6.25FEE1480 pKa = 4.52RR1481 pKa = 11.84GEE1483 pKa = 4.08NTKK1486 pKa = 9.6TPIQKK1491 pKa = 9.21HH1492 pKa = 4.25WRR1494 pKa = 11.84PTVLTEE1500 pKa = 4.11GPPVKK1505 pKa = 10.12IRR1507 pKa = 11.84IEE1509 pKa = 3.85TGEE1512 pKa = 4.09WEE1514 pKa = 4.34KK1515 pKa = 11.25GWNVLVWGRR1524 pKa = 11.84GYY1526 pKa = 11.12AAVKK1530 pKa = 10.59NRR1532 pKa = 11.84DD1533 pKa = 3.13TDD1535 pKa = 3.61KK1536 pKa = 11.36VIWVPSRR1543 pKa = 11.84KK1544 pKa = 9.39VKK1546 pKa = 10.52PDD1548 pKa = 3.13VTQKK1552 pKa = 11.49DD1553 pKa = 3.91EE1554 pKa = 4.15VTKK1557 pKa = 10.61KK1558 pKa = 10.9DD1559 pKa = 3.32EE1560 pKa = 4.5ASPLFAGISDD1570 pKa = 4.51WIPWEE1575 pKa = 4.39DD1576 pKa = 3.79EE1577 pKa = 4.05QEE1579 pKa = 4.24GLQGEE1584 pKa = 4.55TASNKK1589 pKa = 7.35QEE1591 pKa = 4.17RR1592 pKa = 11.84PGEE1595 pKa = 4.05DD1596 pKa = 2.5TLAANEE1602 pKa = 4.27SS1603 pKa = 3.7
Molecular weight: 173.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q04221|VP14_RSVSA Putative trans-acting factor OS=Avian leukosis virus RSA OX=269446 GN=trans-acting factor PE=4 SV=1
MM1 pKa = 7.24EE2 pKa = 4.52AVIRR6 pKa = 11.84EE7 pKa = 4.53GGSLPQVRR15 pKa = 11.84SASRR19 pKa = 11.84NQQRR23 pKa = 11.84SGEE26 pKa = 4.3STKK29 pKa = 10.22GRR31 pKa = 11.84KK32 pKa = 7.66WEE34 pKa = 3.93KK35 pKa = 9.27QLRR38 pKa = 11.84SEE40 pKa = 3.86MRR42 pKa = 11.84RR43 pKa = 11.84WRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 8.69WPHH51 pKa = 6.44LKK53 pKa = 10.13PLAHH57 pKa = 6.81PAISAEE63 pKa = 3.93QLLAVIAPQPRR74 pKa = 11.84PLLLLMWGVVCILPWRR90 pKa = 11.84GWEE93 pKa = 3.84SSRR96 pKa = 11.84ARR98 pKa = 11.84GVTHH102 pKa = 7.13LGGRR106 pKa = 11.84NSQGRR111 pKa = 11.84SQGTRR116 pKa = 11.84VWPLGRR122 pKa = 11.84PP123 pKa = 3.5
MM1 pKa = 7.24EE2 pKa = 4.52AVIRR6 pKa = 11.84EE7 pKa = 4.53GGSLPQVRR15 pKa = 11.84SASRR19 pKa = 11.84NQQRR23 pKa = 11.84SGEE26 pKa = 4.3STKK29 pKa = 10.22GRR31 pKa = 11.84KK32 pKa = 7.66WEE34 pKa = 3.93KK35 pKa = 9.27QLRR38 pKa = 11.84SEE40 pKa = 3.86MRR42 pKa = 11.84RR43 pKa = 11.84WRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 8.69WPHH51 pKa = 6.44LKK53 pKa = 10.13PLAHH57 pKa = 6.81PAISAEE63 pKa = 3.93QLLAVIAPQPRR74 pKa = 11.84PLLLLMWGVVCILPWRR90 pKa = 11.84GWEE93 pKa = 3.84SSRR96 pKa = 11.84ARR98 pKa = 11.84GVTHH102 pKa = 7.13LGGRR106 pKa = 11.84NSQGRR111 pKa = 11.84SQGTRR116 pKa = 11.84VWPLGRR122 pKa = 11.84PP123 pKa = 3.5
Molecular weight: 14.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3033 |
123 |
1603 |
758.3 |
82.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.298 ± 1.032 | 2.176 ± 0.482 |
4.088 ± 0.34 | 5.407 ± 0.394 |
1.912 ± 0.474 | 9.1 ± 0.547 |
1.78 ± 0.209 | 4.946 ± 0.251 |
4.715 ± 0.163 | 10.122 ± 0.413 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.275 ± 0.302 | 2.44 ± 0.399 |
7.451 ± 0.552 | 4.748 ± 0.209 |
6.528 ± 0.768 | 6.759 ± 0.506 |
6.067 ± 0.336 | 6.429 ± 0.294 |
2.374 ± 0.301 | 1.385 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
