
Rice dwarf virus (isolate Fujian) (RDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Phytoreovirus; Rice dwarf virus
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
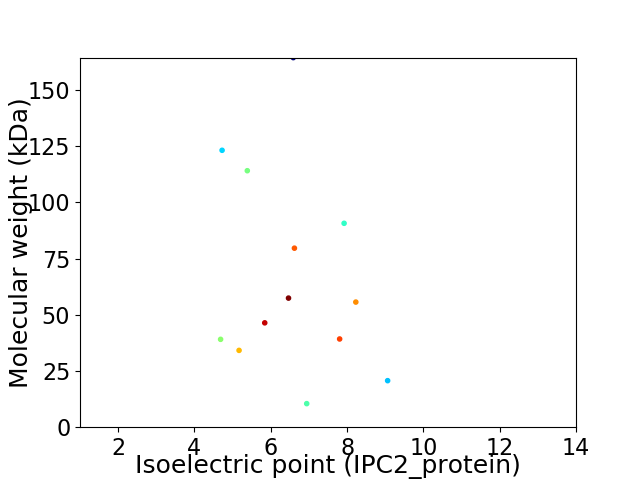
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
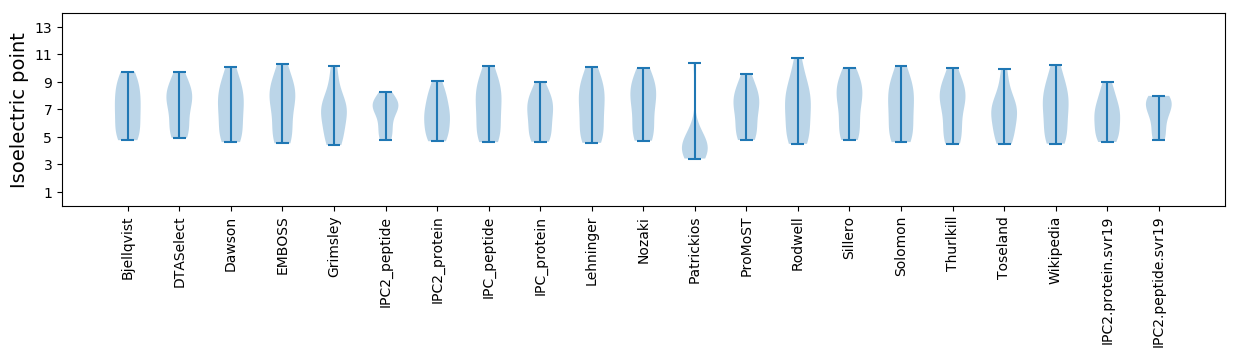
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q98632|P2_RDVF Minor outer capsid protein P2 OS=Rice dwarf virus (isolate Fujian) OX=142804 PE=1 SV=1
MM1 pKa = 8.06AYY3 pKa = 9.78PNDD6 pKa = 3.48VRR8 pKa = 11.84NVWDD12 pKa = 3.78VYY14 pKa = 11.35NVFRR18 pKa = 11.84DD19 pKa = 3.82VPNRR23 pKa = 11.84EE24 pKa = 3.59HH25 pKa = 7.84LIRR28 pKa = 11.84DD29 pKa = 3.22IRR31 pKa = 11.84NGLVTVRR38 pKa = 11.84NLTNMLTNMEE48 pKa = 4.97RR49 pKa = 11.84DD50 pKa = 3.62DD51 pKa = 3.95QLIIAQLSNMMKK63 pKa = 9.98SLSIGIEE70 pKa = 3.81KK71 pKa = 10.07AQNEE75 pKa = 4.15LSKK78 pKa = 11.26LKK80 pKa = 8.63TTDD83 pKa = 3.04ADD85 pKa = 4.12RR86 pKa = 11.84AAVLADD92 pKa = 3.5YY93 pKa = 7.51QTSVLNIEE101 pKa = 4.58RR102 pKa = 11.84NTMLLTGYY110 pKa = 9.84FKK112 pKa = 10.82QLVLDD117 pKa = 3.58LTGYY121 pKa = 10.14VGASVYY127 pKa = 10.53PILPFMITGDD137 pKa = 3.39QSMMVDD143 pKa = 4.15SINVNMKK150 pKa = 10.24NVFDD154 pKa = 5.12DD155 pKa = 3.32KK156 pKa = 11.34HH157 pKa = 5.42EE158 pKa = 4.03QEE160 pKa = 4.58IVLPIHH166 pKa = 6.21PACFVSTITEE176 pKa = 4.12DD177 pKa = 3.39TSSVVYY183 pKa = 10.89ADD185 pKa = 3.47GDD187 pKa = 3.69EE188 pKa = 4.68LYY190 pKa = 10.37SVHH193 pKa = 6.11VRR195 pKa = 11.84HH196 pKa = 6.55EE197 pKa = 4.05NMTMYY202 pKa = 10.86VNVLGEE208 pKa = 4.35TVEE211 pKa = 4.25TRR213 pKa = 11.84QLSMIGEE220 pKa = 4.59SIVPDD225 pKa = 3.91DD226 pKa = 4.5FAPSLLILRR235 pKa = 11.84FSQDD239 pKa = 2.68SVGEE243 pKa = 4.14VFYY246 pKa = 11.0LSHH249 pKa = 7.75DD250 pKa = 4.18NIKK253 pKa = 10.69KK254 pKa = 10.09FLGHH258 pKa = 5.74SLEE261 pKa = 4.22YY262 pKa = 8.67TDD264 pKa = 4.92KK265 pKa = 11.39YY266 pKa = 11.25VIFDD270 pKa = 3.32VARR273 pKa = 11.84RR274 pKa = 11.84ASTTRR279 pKa = 11.84NTITDD284 pKa = 4.27GFCSVDD290 pKa = 3.32GVPYY294 pKa = 10.62LDD296 pKa = 4.4GRR298 pKa = 11.84FIYY301 pKa = 10.25QPSGISADD309 pKa = 3.82SNICAIYY316 pKa = 10.26NSYY319 pKa = 10.73VLDD322 pKa = 3.41VLRR325 pKa = 11.84YY326 pKa = 7.78ITEE329 pKa = 4.31CEE331 pKa = 3.69VDD333 pKa = 3.73TLRR336 pKa = 11.84SVYY339 pKa = 10.55DD340 pKa = 3.59RR341 pKa = 11.84TSSTVFSKK349 pKa = 10.18TDD351 pKa = 3.16VLRR354 pKa = 11.84PRR356 pKa = 11.84LLTMQSNISALSAATPQLANDD377 pKa = 4.76VITSDD382 pKa = 3.73STDD385 pKa = 3.75LLSLGTVLTVSNEE398 pKa = 3.64FTADD402 pKa = 3.48DD403 pKa = 4.06TTLSTSLAGHH413 pKa = 5.89CQVDD417 pKa = 3.95YY418 pKa = 11.68SEE420 pKa = 5.98GSPQDD425 pKa = 3.09KK426 pKa = 10.82SMSIPVSCDD435 pKa = 2.78SSQLASSTVHH445 pKa = 6.47SYY447 pKa = 10.83SADD450 pKa = 3.24ILGHH454 pKa = 5.18GLKK457 pKa = 10.35GDD459 pKa = 3.77RR460 pKa = 11.84NLNLMINVPGLMNPQKK476 pKa = 9.82VTVDD480 pKa = 3.32YY481 pKa = 10.95VYY483 pKa = 11.53SDD485 pKa = 3.74GYY487 pKa = 10.55KK488 pKa = 10.66LNFASVVAPDD498 pKa = 3.21APFWINATLQLSLSPSAHH516 pKa = 5.67NMLSKK521 pKa = 9.92LTPLDD526 pKa = 3.66NDD528 pKa = 3.62ACPGLKK534 pKa = 10.06AQANTPVLVSMTINLDD550 pKa = 3.46DD551 pKa = 4.18ATPALGGEE559 pKa = 4.59VIQNCVFKK567 pKa = 10.46IHH569 pKa = 6.9HH570 pKa = 6.82GDD572 pKa = 3.39DD573 pKa = 3.57VYY575 pKa = 12.0SFVTDD580 pKa = 4.06FDD582 pKa = 4.76VISYY586 pKa = 8.94TSTSGTNCLKK596 pKa = 10.73LISSVDD602 pKa = 3.1ITSQLPSDD610 pKa = 3.15MWIYY614 pKa = 11.16VMNGSPDD621 pKa = 3.21AAFISGDD628 pKa = 3.91SVNMSSVDD636 pKa = 3.1WHH638 pKa = 6.24QSTSQTVGNYY648 pKa = 8.83VYY650 pKa = 10.54ATMKK654 pKa = 10.48AYY656 pKa = 9.75WNVTSYY662 pKa = 11.27DD663 pKa = 3.5VEE665 pKa = 3.95ARR667 pKa = 11.84PYY669 pKa = 8.52ATYY672 pKa = 10.98VPGKK676 pKa = 10.31INFTAIDD683 pKa = 3.74HH684 pKa = 6.99ADD686 pKa = 3.68VFVDD690 pKa = 4.98DD691 pKa = 4.87YY692 pKa = 10.77NTGVNSYY699 pKa = 10.62VIVNSRR705 pKa = 11.84IYY707 pKa = 10.86YY708 pKa = 9.63KK709 pKa = 10.7GNSSIYY715 pKa = 8.71EE716 pKa = 4.1VPSGSFIKK724 pKa = 10.55VSYY727 pKa = 8.03FTSPLKK733 pKa = 11.03NPTVDD738 pKa = 3.4AYY740 pKa = 11.05NAEE743 pKa = 4.07ISRR746 pKa = 11.84NSAYY750 pKa = 10.6LIKK753 pKa = 10.85ANASLDD759 pKa = 3.63SVAAMLSNISNRR771 pKa = 11.84IDD773 pKa = 3.08AMEE776 pKa = 4.43RR777 pKa = 11.84LMEE780 pKa = 4.0PTRR783 pKa = 11.84AHH785 pKa = 7.2RR786 pKa = 11.84IAGVVSSIGGVISLGMPLLGAIVVTIGSIISIADD820 pKa = 3.54PDD822 pKa = 3.85KK823 pKa = 11.39QGIDD827 pKa = 3.48YY828 pKa = 10.63HH829 pKa = 7.88SVANAFMSWCQYY841 pKa = 11.16AAVCRR846 pKa = 11.84YY847 pKa = 9.64EE848 pKa = 4.02YY849 pKa = 11.43GLLKK853 pKa = 10.76RR854 pKa = 11.84GDD856 pKa = 3.77EE857 pKa = 4.2KK858 pKa = 11.55LDD860 pKa = 3.4VLSFMPKK867 pKa = 9.56RR868 pKa = 11.84VVSDD872 pKa = 3.97FKK874 pKa = 11.27NKK876 pKa = 9.57PDD878 pKa = 3.98VISLPEE884 pKa = 4.37LGEE887 pKa = 4.2SVLRR891 pKa = 11.84GSSTDD896 pKa = 3.6YY897 pKa = 11.38LDD899 pKa = 3.42TEE901 pKa = 4.34INIIYY906 pKa = 10.48NDD908 pKa = 3.89MQLLGQGKK916 pKa = 10.1LSDD919 pKa = 3.84WLNKK923 pKa = 8.73TVSKK927 pKa = 11.09VEE929 pKa = 3.79NNAANFFEE937 pKa = 4.65RR938 pKa = 11.84NLVKK942 pKa = 10.66SLANKK947 pKa = 9.36EE948 pKa = 4.18VLPVHH953 pKa = 6.2ARR955 pKa = 11.84VEE957 pKa = 4.03ITQTEE962 pKa = 4.54KK963 pKa = 10.62IGDD966 pKa = 3.84VYY968 pKa = 10.26RR969 pKa = 11.84TTILYY974 pKa = 9.13TGINEE979 pKa = 4.1GSYY982 pKa = 10.76LGGDD986 pKa = 3.69VFASRR991 pKa = 11.84LGDD994 pKa = 3.7KK995 pKa = 10.79NILRR999 pKa = 11.84MNGFEE1004 pKa = 4.22SGPGRR1009 pKa = 11.84FKK1011 pKa = 11.1AIVEE1015 pKa = 4.43STTEE1019 pKa = 3.66VGNFRR1024 pKa = 11.84VVDD1027 pKa = 3.54WTVSGMSRR1035 pKa = 11.84YY1036 pKa = 9.32EE1037 pKa = 3.54IYY1039 pKa = 10.65AAAGEE1044 pKa = 4.64VYY1046 pKa = 9.95PSKK1049 pKa = 10.86DD1050 pKa = 3.21PSHH1053 pKa = 7.29ADD1055 pKa = 3.16VQLLYY1060 pKa = 10.83EE1061 pKa = 4.53SIVRR1065 pKa = 11.84DD1066 pKa = 3.39LTTRR1070 pKa = 11.84DD1071 pKa = 2.98GSFVLKK1077 pKa = 10.58HH1078 pKa = 6.41HH1079 pKa = 7.34DD1080 pKa = 3.65VLLLPGQLDD1089 pKa = 3.5AFEE1092 pKa = 4.37EE1093 pKa = 4.73LIIKK1097 pKa = 9.07NASNYY1102 pKa = 9.08QYY1104 pKa = 11.81AFIGSNCSKK1113 pKa = 10.84LCAA1116 pKa = 4.26
MM1 pKa = 8.06AYY3 pKa = 9.78PNDD6 pKa = 3.48VRR8 pKa = 11.84NVWDD12 pKa = 3.78VYY14 pKa = 11.35NVFRR18 pKa = 11.84DD19 pKa = 3.82VPNRR23 pKa = 11.84EE24 pKa = 3.59HH25 pKa = 7.84LIRR28 pKa = 11.84DD29 pKa = 3.22IRR31 pKa = 11.84NGLVTVRR38 pKa = 11.84NLTNMLTNMEE48 pKa = 4.97RR49 pKa = 11.84DD50 pKa = 3.62DD51 pKa = 3.95QLIIAQLSNMMKK63 pKa = 9.98SLSIGIEE70 pKa = 3.81KK71 pKa = 10.07AQNEE75 pKa = 4.15LSKK78 pKa = 11.26LKK80 pKa = 8.63TTDD83 pKa = 3.04ADD85 pKa = 4.12RR86 pKa = 11.84AAVLADD92 pKa = 3.5YY93 pKa = 7.51QTSVLNIEE101 pKa = 4.58RR102 pKa = 11.84NTMLLTGYY110 pKa = 9.84FKK112 pKa = 10.82QLVLDD117 pKa = 3.58LTGYY121 pKa = 10.14VGASVYY127 pKa = 10.53PILPFMITGDD137 pKa = 3.39QSMMVDD143 pKa = 4.15SINVNMKK150 pKa = 10.24NVFDD154 pKa = 5.12DD155 pKa = 3.32KK156 pKa = 11.34HH157 pKa = 5.42EE158 pKa = 4.03QEE160 pKa = 4.58IVLPIHH166 pKa = 6.21PACFVSTITEE176 pKa = 4.12DD177 pKa = 3.39TSSVVYY183 pKa = 10.89ADD185 pKa = 3.47GDD187 pKa = 3.69EE188 pKa = 4.68LYY190 pKa = 10.37SVHH193 pKa = 6.11VRR195 pKa = 11.84HH196 pKa = 6.55EE197 pKa = 4.05NMTMYY202 pKa = 10.86VNVLGEE208 pKa = 4.35TVEE211 pKa = 4.25TRR213 pKa = 11.84QLSMIGEE220 pKa = 4.59SIVPDD225 pKa = 3.91DD226 pKa = 4.5FAPSLLILRR235 pKa = 11.84FSQDD239 pKa = 2.68SVGEE243 pKa = 4.14VFYY246 pKa = 11.0LSHH249 pKa = 7.75DD250 pKa = 4.18NIKK253 pKa = 10.69KK254 pKa = 10.09FLGHH258 pKa = 5.74SLEE261 pKa = 4.22YY262 pKa = 8.67TDD264 pKa = 4.92KK265 pKa = 11.39YY266 pKa = 11.25VIFDD270 pKa = 3.32VARR273 pKa = 11.84RR274 pKa = 11.84ASTTRR279 pKa = 11.84NTITDD284 pKa = 4.27GFCSVDD290 pKa = 3.32GVPYY294 pKa = 10.62LDD296 pKa = 4.4GRR298 pKa = 11.84FIYY301 pKa = 10.25QPSGISADD309 pKa = 3.82SNICAIYY316 pKa = 10.26NSYY319 pKa = 10.73VLDD322 pKa = 3.41VLRR325 pKa = 11.84YY326 pKa = 7.78ITEE329 pKa = 4.31CEE331 pKa = 3.69VDD333 pKa = 3.73TLRR336 pKa = 11.84SVYY339 pKa = 10.55DD340 pKa = 3.59RR341 pKa = 11.84TSSTVFSKK349 pKa = 10.18TDD351 pKa = 3.16VLRR354 pKa = 11.84PRR356 pKa = 11.84LLTMQSNISALSAATPQLANDD377 pKa = 4.76VITSDD382 pKa = 3.73STDD385 pKa = 3.75LLSLGTVLTVSNEE398 pKa = 3.64FTADD402 pKa = 3.48DD403 pKa = 4.06TTLSTSLAGHH413 pKa = 5.89CQVDD417 pKa = 3.95YY418 pKa = 11.68SEE420 pKa = 5.98GSPQDD425 pKa = 3.09KK426 pKa = 10.82SMSIPVSCDD435 pKa = 2.78SSQLASSTVHH445 pKa = 6.47SYY447 pKa = 10.83SADD450 pKa = 3.24ILGHH454 pKa = 5.18GLKK457 pKa = 10.35GDD459 pKa = 3.77RR460 pKa = 11.84NLNLMINVPGLMNPQKK476 pKa = 9.82VTVDD480 pKa = 3.32YY481 pKa = 10.95VYY483 pKa = 11.53SDD485 pKa = 3.74GYY487 pKa = 10.55KK488 pKa = 10.66LNFASVVAPDD498 pKa = 3.21APFWINATLQLSLSPSAHH516 pKa = 5.67NMLSKK521 pKa = 9.92LTPLDD526 pKa = 3.66NDD528 pKa = 3.62ACPGLKK534 pKa = 10.06AQANTPVLVSMTINLDD550 pKa = 3.46DD551 pKa = 4.18ATPALGGEE559 pKa = 4.59VIQNCVFKK567 pKa = 10.46IHH569 pKa = 6.9HH570 pKa = 6.82GDD572 pKa = 3.39DD573 pKa = 3.57VYY575 pKa = 12.0SFVTDD580 pKa = 4.06FDD582 pKa = 4.76VISYY586 pKa = 8.94TSTSGTNCLKK596 pKa = 10.73LISSVDD602 pKa = 3.1ITSQLPSDD610 pKa = 3.15MWIYY614 pKa = 11.16VMNGSPDD621 pKa = 3.21AAFISGDD628 pKa = 3.91SVNMSSVDD636 pKa = 3.1WHH638 pKa = 6.24QSTSQTVGNYY648 pKa = 8.83VYY650 pKa = 10.54ATMKK654 pKa = 10.48AYY656 pKa = 9.75WNVTSYY662 pKa = 11.27DD663 pKa = 3.5VEE665 pKa = 3.95ARR667 pKa = 11.84PYY669 pKa = 8.52ATYY672 pKa = 10.98VPGKK676 pKa = 10.31INFTAIDD683 pKa = 3.74HH684 pKa = 6.99ADD686 pKa = 3.68VFVDD690 pKa = 4.98DD691 pKa = 4.87YY692 pKa = 10.77NTGVNSYY699 pKa = 10.62VIVNSRR705 pKa = 11.84IYY707 pKa = 10.86YY708 pKa = 9.63KK709 pKa = 10.7GNSSIYY715 pKa = 8.71EE716 pKa = 4.1VPSGSFIKK724 pKa = 10.55VSYY727 pKa = 8.03FTSPLKK733 pKa = 11.03NPTVDD738 pKa = 3.4AYY740 pKa = 11.05NAEE743 pKa = 4.07ISRR746 pKa = 11.84NSAYY750 pKa = 10.6LIKK753 pKa = 10.85ANASLDD759 pKa = 3.63SVAAMLSNISNRR771 pKa = 11.84IDD773 pKa = 3.08AMEE776 pKa = 4.43RR777 pKa = 11.84LMEE780 pKa = 4.0PTRR783 pKa = 11.84AHH785 pKa = 7.2RR786 pKa = 11.84IAGVVSSIGGVISLGMPLLGAIVVTIGSIISIADD820 pKa = 3.54PDD822 pKa = 3.85KK823 pKa = 11.39QGIDD827 pKa = 3.48YY828 pKa = 10.63HH829 pKa = 7.88SVANAFMSWCQYY841 pKa = 11.16AAVCRR846 pKa = 11.84YY847 pKa = 9.64EE848 pKa = 4.02YY849 pKa = 11.43GLLKK853 pKa = 10.76RR854 pKa = 11.84GDD856 pKa = 3.77EE857 pKa = 4.2KK858 pKa = 11.55LDD860 pKa = 3.4VLSFMPKK867 pKa = 9.56RR868 pKa = 11.84VVSDD872 pKa = 3.97FKK874 pKa = 11.27NKK876 pKa = 9.57PDD878 pKa = 3.98VISLPEE884 pKa = 4.37LGEE887 pKa = 4.2SVLRR891 pKa = 11.84GSSTDD896 pKa = 3.6YY897 pKa = 11.38LDD899 pKa = 3.42TEE901 pKa = 4.34INIIYY906 pKa = 10.48NDD908 pKa = 3.89MQLLGQGKK916 pKa = 10.1LSDD919 pKa = 3.84WLNKK923 pKa = 8.73TVSKK927 pKa = 11.09VEE929 pKa = 3.79NNAANFFEE937 pKa = 4.65RR938 pKa = 11.84NLVKK942 pKa = 10.66SLANKK947 pKa = 9.36EE948 pKa = 4.18VLPVHH953 pKa = 6.2ARR955 pKa = 11.84VEE957 pKa = 4.03ITQTEE962 pKa = 4.54KK963 pKa = 10.62IGDD966 pKa = 3.84VYY968 pKa = 10.26RR969 pKa = 11.84TTILYY974 pKa = 9.13TGINEE979 pKa = 4.1GSYY982 pKa = 10.76LGGDD986 pKa = 3.69VFASRR991 pKa = 11.84LGDD994 pKa = 3.7KK995 pKa = 10.79NILRR999 pKa = 11.84MNGFEE1004 pKa = 4.22SGPGRR1009 pKa = 11.84FKK1011 pKa = 11.1AIVEE1015 pKa = 4.43STTEE1019 pKa = 3.66VGNFRR1024 pKa = 11.84VVDD1027 pKa = 3.54WTVSGMSRR1035 pKa = 11.84YY1036 pKa = 9.32EE1037 pKa = 3.54IYY1039 pKa = 10.65AAAGEE1044 pKa = 4.64VYY1046 pKa = 9.95PSKK1049 pKa = 10.86DD1050 pKa = 3.21PSHH1053 pKa = 7.29ADD1055 pKa = 3.16VQLLYY1060 pKa = 10.83EE1061 pKa = 4.53SIVRR1065 pKa = 11.84DD1066 pKa = 3.39LTTRR1070 pKa = 11.84DD1071 pKa = 2.98GSFVLKK1077 pKa = 10.58HH1078 pKa = 6.41HH1079 pKa = 7.34DD1080 pKa = 3.65VLLLPGQLDD1089 pKa = 3.5AFEE1092 pKa = 4.37EE1093 pKa = 4.73LIIKK1097 pKa = 9.07NASNYY1102 pKa = 9.08QYY1104 pKa = 11.81AFIGSNCSKK1113 pKa = 10.84LCAA1116 pKa = 4.26
Molecular weight: 123.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q85443|NSP12_RDVF Non-structural protein 12A OS=Rice dwarf virus (isolate Fujian) OX=142804 PE=2 SV=1
MM1 pKa = 7.79SGTLPLAMTASEE13 pKa = 4.89SFVGMQVLAQDD24 pKa = 4.45KK25 pKa = 9.24EE26 pKa = 4.54VKK28 pKa = 9.37ATFIALDD35 pKa = 3.48RR36 pKa = 11.84KK37 pKa = 10.48LPANLKK43 pKa = 9.22VPYY46 pKa = 8.92MKK48 pKa = 10.12NAKK51 pKa = 9.48YY52 pKa = 8.07RR53 pKa = 11.84TCICPSSNHH62 pKa = 6.32LVDD65 pKa = 5.24DD66 pKa = 4.75CVCEE70 pKa = 4.06DD71 pKa = 4.05VIIAYY76 pKa = 7.41TAHH79 pKa = 7.1RR80 pKa = 11.84NNAVAALLYY89 pKa = 10.55SDD91 pKa = 4.73GNVIHH96 pKa = 7.52RR97 pKa = 11.84SGTLKK102 pKa = 10.56PKK104 pKa = 9.33SQNRR108 pKa = 11.84FDD110 pKa = 4.07LRR112 pKa = 11.84GFLTSVNPGEE122 pKa = 4.24SSKK125 pKa = 11.68AEE127 pKa = 4.1AGTSKK132 pKa = 9.31STQKK136 pKa = 11.05AYY138 pKa = 10.49DD139 pKa = 3.85RR140 pKa = 11.84KK141 pKa = 10.7DD142 pKa = 3.14KK143 pKa = 11.35SPSKK147 pKa = 10.79GKK149 pKa = 10.02NSKK152 pKa = 10.37KK153 pKa = 10.24GGKK156 pKa = 9.3KK157 pKa = 9.86SSSEE161 pKa = 4.1RR162 pKa = 11.84KK163 pKa = 8.84RR164 pKa = 11.84KK165 pKa = 9.4EE166 pKa = 3.57YY167 pKa = 10.81SSNSEE172 pKa = 3.84TDD174 pKa = 3.3LSSDD178 pKa = 2.88SDD180 pKa = 3.99ANTRR184 pKa = 11.84KK185 pKa = 9.88SKK187 pKa = 10.83RR188 pKa = 11.84KK189 pKa = 9.37
MM1 pKa = 7.79SGTLPLAMTASEE13 pKa = 4.89SFVGMQVLAQDD24 pKa = 4.45KK25 pKa = 9.24EE26 pKa = 4.54VKK28 pKa = 9.37ATFIALDD35 pKa = 3.48RR36 pKa = 11.84KK37 pKa = 10.48LPANLKK43 pKa = 9.22VPYY46 pKa = 8.92MKK48 pKa = 10.12NAKK51 pKa = 9.48YY52 pKa = 8.07RR53 pKa = 11.84TCICPSSNHH62 pKa = 6.32LVDD65 pKa = 5.24DD66 pKa = 4.75CVCEE70 pKa = 4.06DD71 pKa = 4.05VIIAYY76 pKa = 7.41TAHH79 pKa = 7.1RR80 pKa = 11.84NNAVAALLYY89 pKa = 10.55SDD91 pKa = 4.73GNVIHH96 pKa = 7.52RR97 pKa = 11.84SGTLKK102 pKa = 10.56PKK104 pKa = 9.33SQNRR108 pKa = 11.84FDD110 pKa = 4.07LRR112 pKa = 11.84GFLTSVNPGEE122 pKa = 4.24SSKK125 pKa = 11.68AEE127 pKa = 4.1AGTSKK132 pKa = 9.31STQKK136 pKa = 11.05AYY138 pKa = 10.49DD139 pKa = 3.85RR140 pKa = 11.84KK141 pKa = 10.7DD142 pKa = 3.14KK143 pKa = 11.35SPSKK147 pKa = 10.79GKK149 pKa = 10.02NSKK152 pKa = 10.37KK153 pKa = 10.24GGKK156 pKa = 9.3KK157 pKa = 9.86SSSEE161 pKa = 4.1RR162 pKa = 11.84KK163 pKa = 8.84RR164 pKa = 11.84KK165 pKa = 9.4EE166 pKa = 3.57YY167 pKa = 10.81SSNSEE172 pKa = 3.84TDD174 pKa = 3.3LSSDD178 pKa = 2.88SDD180 pKa = 3.99ANTRR184 pKa = 11.84KK185 pKa = 9.88SKK187 pKa = 10.83RR188 pKa = 11.84KK189 pKa = 9.37
Molecular weight: 20.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7840 |
92 |
1444 |
603.1 |
67.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.722 ± 0.312 | 1.161 ± 0.117 |
6.314 ± 0.314 | 4.579 ± 0.286 |
4.26 ± 0.243 | 5.37 ± 0.261 |
2.092 ± 0.164 | 6.237 ± 0.218 |
5.255 ± 0.474 | 9.592 ± 0.236 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.908 ± 0.112 | 5.357 ± 0.179 |
4.26 ± 0.235 | 3.048 ± 0.181 |
4.898 ± 0.257 | 9.63 ± 0.388 |
6.173 ± 0.301 | 7.258 ± 0.459 |
0.842 ± 0.138 | 4.043 ± 0.302 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
