
Luna mammarenavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Arenaviridae; Mammarenavirus
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
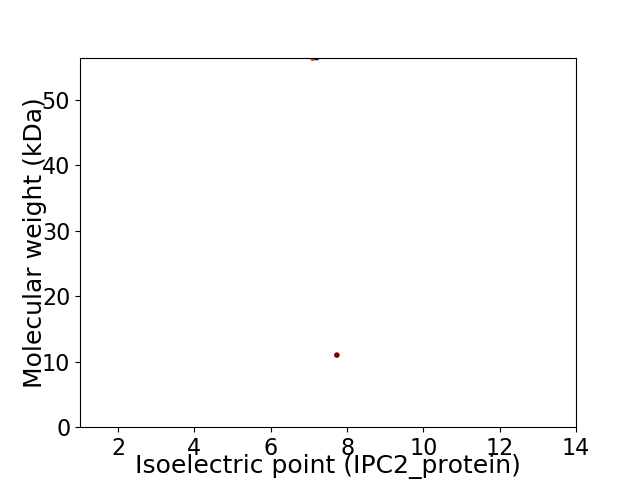
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4VYZ4|A0A0U4VYZ4_9VIRU Pre-glycoprotein polyprotein GP complex OS=Luna mammarenavirus OX=883876 GN=GP PE=3 SV=1
MM1 pKa = 6.88GQIVTFFQEE10 pKa = 4.01VPHH13 pKa = 8.29IIEE16 pKa = 3.78EE17 pKa = 4.17VMNIVLITLSLLAILKK33 pKa = 9.82GVYY36 pKa = 9.96NLATCGLLGLISFLFLCGRR55 pKa = 11.84SCSITVKK62 pKa = 9.85DD63 pKa = 3.69TYY65 pKa = 9.48EE66 pKa = 3.98LRR68 pKa = 11.84SVEE71 pKa = 4.79LDD73 pKa = 3.15MSTLNYY79 pKa = 9.69TMPLSCSKK87 pKa = 10.84NNSHH91 pKa = 7.33HH92 pKa = 6.6YY93 pKa = 9.98IQTFNDD99 pKa = 3.26TGLEE103 pKa = 4.01LTLTNDD109 pKa = 2.91SLLNHH114 pKa = 6.9KK115 pKa = 9.74FCNLSDD121 pKa = 3.19AHH123 pKa = 6.64KK124 pKa = 10.65RR125 pKa = 11.84NLYY128 pKa = 10.46DD129 pKa = 3.31HH130 pKa = 6.95TLMSVITTFHH140 pKa = 6.68LSIPNFNQYY149 pKa = 9.17EE150 pKa = 4.06AMACDD155 pKa = 4.1FNGGKK160 pKa = 8.12ITVQYY165 pKa = 9.95NLSHH169 pKa = 6.86SSPVDD174 pKa = 3.29AANHH178 pKa = 6.36CGTVANGVLDD188 pKa = 3.82TFRR191 pKa = 11.84RR192 pKa = 11.84MHH194 pKa = 7.04WGPAHH199 pKa = 6.6FPDD202 pKa = 4.92PPFQIGSVNCIQTSYY217 pKa = 10.99KK218 pKa = 10.4YY219 pKa = 10.93LIIQNTTWEE228 pKa = 4.48DD229 pKa = 3.31HH230 pKa = 6.37CQMSRR235 pKa = 11.84PTPMGYY241 pKa = 10.6LSLIAQRR248 pKa = 11.84TRR250 pKa = 11.84QLYY253 pKa = 9.02ISRR256 pKa = 11.84RR257 pKa = 11.84LMGTFTWTLSDD268 pKa = 3.41SAGNNLPGGYY278 pKa = 9.76CLQRR282 pKa = 11.84WMLIEE287 pKa = 6.12AEE289 pKa = 4.5MKK291 pKa = 10.75CFGNTAVAKK300 pKa = 10.44CNQQHH305 pKa = 6.86DD306 pKa = 4.37EE307 pKa = 4.55EE308 pKa = 5.15FCDD311 pKa = 3.62MLRR314 pKa = 11.84LFDD317 pKa = 4.54FNKK320 pKa = 10.18EE321 pKa = 3.9AINRR325 pKa = 11.84LRR327 pKa = 11.84VEE329 pKa = 4.12AEE331 pKa = 3.66KK332 pKa = 10.9SINLINKK339 pKa = 8.44AVNSLINDD347 pKa = 3.3QLIIRR352 pKa = 11.84NHH354 pKa = 6.02LRR356 pKa = 11.84DD357 pKa = 3.78LMGIPYY363 pKa = 10.31CNYY366 pKa = 10.46SKK368 pKa = 10.66FWYY371 pKa = 10.77LNDD374 pKa = 3.4TRR376 pKa = 11.84SGRR379 pKa = 11.84TSLPKK384 pKa = 9.92CWKK387 pKa = 9.79VSNNSYY393 pKa = 10.95LNEE396 pKa = 3.99THH398 pKa = 7.01FSDD401 pKa = 5.43EE402 pKa = 4.38IEE404 pKa = 4.17EE405 pKa = 4.23EE406 pKa = 4.26ANNMITEE413 pKa = 4.45MLRR416 pKa = 11.84KK417 pKa = 9.39EE418 pKa = 4.04YY419 pKa = 10.44EE420 pKa = 4.15KK421 pKa = 10.87RR422 pKa = 11.84QSTTPLGLVDD432 pKa = 5.27LFVFSTSFYY441 pKa = 10.09LISIFLHH448 pKa = 6.43LIKK451 pKa = 10.35IPTHH455 pKa = 4.51RR456 pKa = 11.84HH457 pKa = 4.11IVGKK461 pKa = 10.3GCPKK465 pKa = 9.68PHH467 pKa = 7.33RR468 pKa = 11.84LNHH471 pKa = 5.72MAICSCGLYY480 pKa = 9.48KK481 pKa = 10.54QPGLPVRR488 pKa = 11.84WKK490 pKa = 10.28RR491 pKa = 3.49
MM1 pKa = 6.88GQIVTFFQEE10 pKa = 4.01VPHH13 pKa = 8.29IIEE16 pKa = 3.78EE17 pKa = 4.17VMNIVLITLSLLAILKK33 pKa = 9.82GVYY36 pKa = 9.96NLATCGLLGLISFLFLCGRR55 pKa = 11.84SCSITVKK62 pKa = 9.85DD63 pKa = 3.69TYY65 pKa = 9.48EE66 pKa = 3.98LRR68 pKa = 11.84SVEE71 pKa = 4.79LDD73 pKa = 3.15MSTLNYY79 pKa = 9.69TMPLSCSKK87 pKa = 10.84NNSHH91 pKa = 7.33HH92 pKa = 6.6YY93 pKa = 9.98IQTFNDD99 pKa = 3.26TGLEE103 pKa = 4.01LTLTNDD109 pKa = 2.91SLLNHH114 pKa = 6.9KK115 pKa = 9.74FCNLSDD121 pKa = 3.19AHH123 pKa = 6.64KK124 pKa = 10.65RR125 pKa = 11.84NLYY128 pKa = 10.46DD129 pKa = 3.31HH130 pKa = 6.95TLMSVITTFHH140 pKa = 6.68LSIPNFNQYY149 pKa = 9.17EE150 pKa = 4.06AMACDD155 pKa = 4.1FNGGKK160 pKa = 8.12ITVQYY165 pKa = 9.95NLSHH169 pKa = 6.86SSPVDD174 pKa = 3.29AANHH178 pKa = 6.36CGTVANGVLDD188 pKa = 3.82TFRR191 pKa = 11.84RR192 pKa = 11.84MHH194 pKa = 7.04WGPAHH199 pKa = 6.6FPDD202 pKa = 4.92PPFQIGSVNCIQTSYY217 pKa = 10.99KK218 pKa = 10.4YY219 pKa = 10.93LIIQNTTWEE228 pKa = 4.48DD229 pKa = 3.31HH230 pKa = 6.37CQMSRR235 pKa = 11.84PTPMGYY241 pKa = 10.6LSLIAQRR248 pKa = 11.84TRR250 pKa = 11.84QLYY253 pKa = 9.02ISRR256 pKa = 11.84RR257 pKa = 11.84LMGTFTWTLSDD268 pKa = 3.41SAGNNLPGGYY278 pKa = 9.76CLQRR282 pKa = 11.84WMLIEE287 pKa = 6.12AEE289 pKa = 4.5MKK291 pKa = 10.75CFGNTAVAKK300 pKa = 10.44CNQQHH305 pKa = 6.86DD306 pKa = 4.37EE307 pKa = 4.55EE308 pKa = 5.15FCDD311 pKa = 3.62MLRR314 pKa = 11.84LFDD317 pKa = 4.54FNKK320 pKa = 10.18EE321 pKa = 3.9AINRR325 pKa = 11.84LRR327 pKa = 11.84VEE329 pKa = 4.12AEE331 pKa = 3.66KK332 pKa = 10.9SINLINKK339 pKa = 8.44AVNSLINDD347 pKa = 3.3QLIIRR352 pKa = 11.84NHH354 pKa = 6.02LRR356 pKa = 11.84DD357 pKa = 3.78LMGIPYY363 pKa = 10.31CNYY366 pKa = 10.46SKK368 pKa = 10.66FWYY371 pKa = 10.77LNDD374 pKa = 3.4TRR376 pKa = 11.84SGRR379 pKa = 11.84TSLPKK384 pKa = 9.92CWKK387 pKa = 9.79VSNNSYY393 pKa = 10.95LNEE396 pKa = 3.99THH398 pKa = 7.01FSDD401 pKa = 5.43EE402 pKa = 4.38IEE404 pKa = 4.17EE405 pKa = 4.23EE406 pKa = 4.26ANNMITEE413 pKa = 4.45MLRR416 pKa = 11.84KK417 pKa = 9.39EE418 pKa = 4.04YY419 pKa = 10.44EE420 pKa = 4.15KK421 pKa = 10.87RR422 pKa = 11.84QSTTPLGLVDD432 pKa = 5.27LFVFSTSFYY441 pKa = 10.09LISIFLHH448 pKa = 6.43LIKK451 pKa = 10.35IPTHH455 pKa = 4.51RR456 pKa = 11.84HH457 pKa = 4.11IVGKK461 pKa = 10.3GCPKK465 pKa = 9.68PHH467 pKa = 7.33RR468 pKa = 11.84LNHH471 pKa = 5.72MAICSCGLYY480 pKa = 9.48KK481 pKa = 10.54QPGLPVRR488 pKa = 11.84WKK490 pKa = 10.28RR491 pKa = 3.49
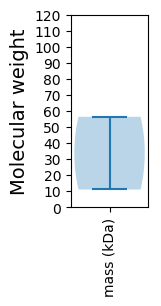
Molecular weight: 56.34 kDa
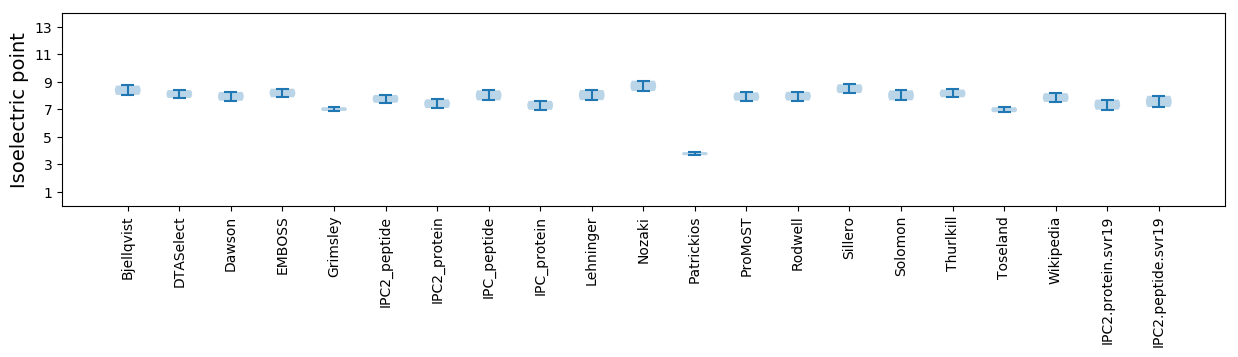
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4VYZ4|A0A0U4VYZ4_9VIRU Pre-glycoprotein polyprotein GP complex OS=Luna mammarenavirus OX=883876 GN=GP PE=3 SV=1
MM1 pKa = 7.74GKK3 pKa = 8.68NQSKK7 pKa = 10.74SNRR10 pKa = 11.84TPDD13 pKa = 3.71NIEE16 pKa = 3.93PRR18 pKa = 11.84STLVPDD24 pKa = 4.45ASGHH28 pKa = 5.26GPEE31 pKa = 4.61FCKK34 pKa = 10.86SCWFEE39 pKa = 3.49RR40 pKa = 11.84RR41 pKa = 11.84GLVKK45 pKa = 10.77CHH47 pKa = 6.37DD48 pKa = 4.86HH49 pKa = 5.75YY50 pKa = 12.07LCMNCLTLLLTISDD64 pKa = 4.05RR65 pKa = 11.84CPICKK70 pKa = 10.12YY71 pKa = 10.07PLPTKK76 pKa = 10.36LQLPKK81 pKa = 10.49TPTAPHH87 pKa = 5.94EE88 pKa = 4.49TAHH91 pKa = 6.63PPPPYY96 pKa = 10.53SPP98 pKa = 4.25
MM1 pKa = 7.74GKK3 pKa = 8.68NQSKK7 pKa = 10.74SNRR10 pKa = 11.84TPDD13 pKa = 3.71NIEE16 pKa = 3.93PRR18 pKa = 11.84STLVPDD24 pKa = 4.45ASGHH28 pKa = 5.26GPEE31 pKa = 4.61FCKK34 pKa = 10.86SCWFEE39 pKa = 3.49RR40 pKa = 11.84RR41 pKa = 11.84GLVKK45 pKa = 10.77CHH47 pKa = 6.37DD48 pKa = 4.86HH49 pKa = 5.75YY50 pKa = 12.07LCMNCLTLLLTISDD64 pKa = 4.05RR65 pKa = 11.84CPICKK70 pKa = 10.12YY71 pKa = 10.07PLPTKK76 pKa = 10.36LQLPKK81 pKa = 10.49TPTAPHH87 pKa = 5.94EE88 pKa = 4.49TAHH91 pKa = 6.63PPPPYY96 pKa = 10.53SPP98 pKa = 4.25
Molecular weight: 11.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
589 |
98 |
491 |
294.5 |
33.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.735 ± 0.293 | 4.244 ± 1.261 |
4.075 ± 0.003 | 4.584 ± 0.219 |
4.244 ± 0.959 | 4.924 ± 0.366 |
4.244 ± 0.373 | 6.282 ± 1.402 |
4.924 ± 0.966 | 11.205 ± 0.436 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.226 ± 0.516 | 6.961 ± 1.253 |
5.772 ± 4.149 | 3.226 ± 0.516 |
4.924 ± 0.078 | 7.301 ± 0.069 |
7.131 ± 0.449 | 3.905 ± 0.811 |
1.358 ± 0.147 | 3.735 ± 0.293 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
