
Lynx canadensis associated microvirus CLP 9413
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
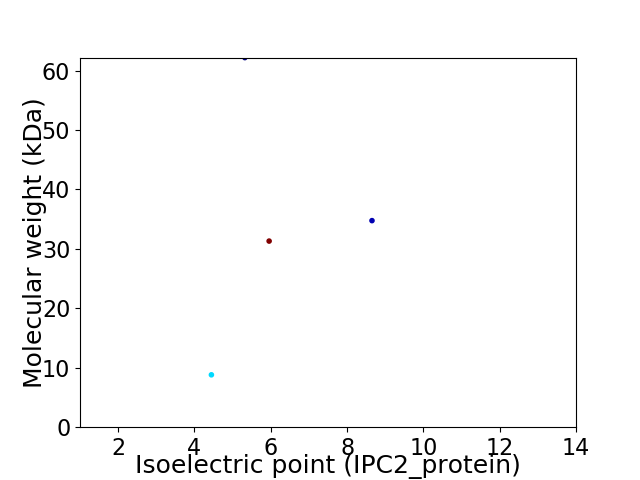
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CI87|A0A2Z5CI87_9VIRU Replication initiation protein OS=Lynx canadensis associated microvirus CLP 9413 OX=2219141 PE=4 SV=1
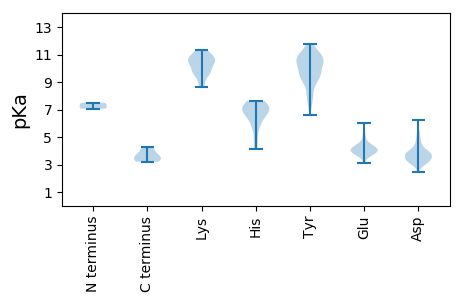
MM1 pKa = 7.39TFGVYY6 pKa = 10.57AIRR9 pKa = 11.84DD10 pKa = 3.7VKK12 pKa = 9.87TGFMSPHH19 pKa = 7.19VEE21 pKa = 3.91EE22 pKa = 5.02SDD24 pKa = 3.43PVAIRR29 pKa = 11.84NFEE32 pKa = 4.26HH33 pKa = 7.11AVQSSDD39 pKa = 4.2GILMSHH45 pKa = 6.93AADD48 pKa = 4.17FSFYY52 pKa = 10.88RR53 pKa = 11.84LGTFDD58 pKa = 3.96SATGLIAADD67 pKa = 3.81TMPTLIMEE75 pKa = 6.05AIDD78 pKa = 3.82CVPP81 pKa = 3.47
MM1 pKa = 7.39TFGVYY6 pKa = 10.57AIRR9 pKa = 11.84DD10 pKa = 3.7VKK12 pKa = 9.87TGFMSPHH19 pKa = 7.19VEE21 pKa = 3.91EE22 pKa = 5.02SDD24 pKa = 3.43PVAIRR29 pKa = 11.84NFEE32 pKa = 4.26HH33 pKa = 7.11AVQSSDD39 pKa = 4.2GILMSHH45 pKa = 6.93AADD48 pKa = 4.17FSFYY52 pKa = 10.88RR53 pKa = 11.84LGTFDD58 pKa = 3.96SATGLIAADD67 pKa = 3.81TMPTLIMEE75 pKa = 6.05AIDD78 pKa = 3.82CVPP81 pKa = 3.47
Molecular weight: 8.82 kDa
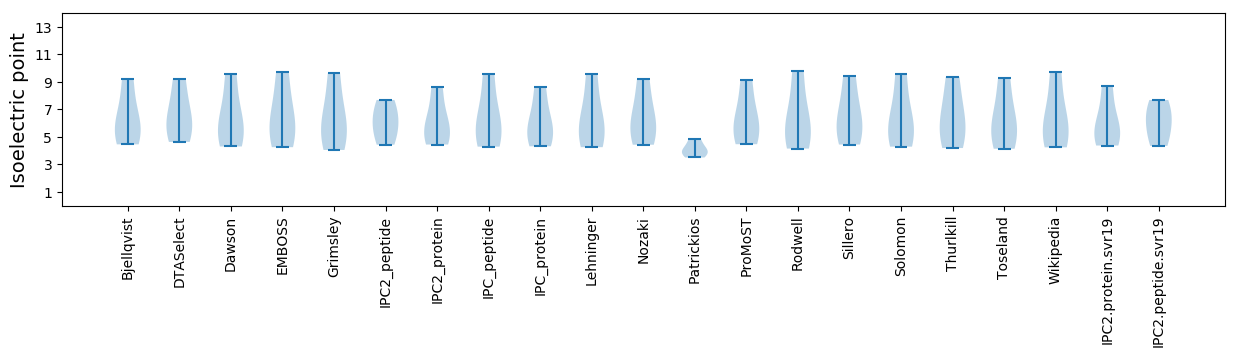
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CHX0|A0A2Z5CHX0_9VIRU Nonstructural protein OS=Lynx canadensis associated microvirus CLP 9413 OX=2219141 PE=4 SV=1
MM1 pKa = 7.5AVNSAANAAQSGVTWQNNPAMSHH24 pKa = 6.0AVSQASGMIDD34 pKa = 4.0KK35 pKa = 10.66ISGIADD41 pKa = 3.72RR42 pKa = 11.84NSAWSAAQAEE52 pKa = 4.47QLRR55 pKa = 11.84DD56 pKa = 3.13WQTQQNKK63 pKa = 9.36IAMDD67 pKa = 3.66YY68 pKa = 10.63NAAEE72 pKa = 4.09AAKK75 pKa = 10.52NRR77 pKa = 11.84DD78 pKa = 2.96WQEE81 pKa = 3.78YY82 pKa = 7.88MSNTAHH88 pKa = 5.58QRR90 pKa = 11.84EE91 pKa = 4.28VRR93 pKa = 11.84DD94 pKa = 3.79LKK96 pKa = 11.22AAGLNPILSAMGGNGAAVGSGATASGVTSAGAKK129 pKa = 9.73GDD131 pKa = 3.66RR132 pKa = 11.84DD133 pKa = 3.56TSANNAIVSVLGSLLNSMTSMAMSTNSAITNLAVADD169 pKa = 4.64KK170 pKa = 9.78YY171 pKa = 11.7NAMSKK176 pKa = 8.76YY177 pKa = 9.08TADD180 pKa = 3.77LSSNTQLAGYY190 pKa = 9.18RR191 pKa = 11.84ISAEE195 pKa = 4.14TALNTANINAAVSRR209 pKa = 11.84YY210 pKa = 9.99VSDD213 pKa = 4.06NNLKK217 pKa = 10.54GSQAMAAATKK227 pKa = 10.37ISAQLHH233 pKa = 6.0ADD235 pKa = 3.32ASKK238 pKa = 9.72YY239 pKa = 10.67AADD242 pKa = 3.88KK243 pKa = 11.21GYY245 pKa = 8.79LTSTDD250 pKa = 3.3VAKK253 pKa = 10.81INADD257 pKa = 2.93INKK260 pKa = 8.89QLKK263 pKa = 9.76QMGIDD268 pKa = 3.42AQFDD272 pKa = 3.6FARR275 pKa = 11.84DD276 pKa = 3.64YY277 pKa = 11.14PSNAFQAVGSISDD290 pKa = 3.74LFGNASGGSGKK301 pKa = 10.5GVSSLFNSVVNGLRR315 pKa = 11.84GGFGEE320 pKa = 4.41RR321 pKa = 11.84KK322 pKa = 9.66SGFSGGSKK330 pKa = 10.15GYY332 pKa = 9.6SGKK335 pKa = 10.44RR336 pKa = 3.21
MM1 pKa = 7.5AVNSAANAAQSGVTWQNNPAMSHH24 pKa = 6.0AVSQASGMIDD34 pKa = 4.0KK35 pKa = 10.66ISGIADD41 pKa = 3.72RR42 pKa = 11.84NSAWSAAQAEE52 pKa = 4.47QLRR55 pKa = 11.84DD56 pKa = 3.13WQTQQNKK63 pKa = 9.36IAMDD67 pKa = 3.66YY68 pKa = 10.63NAAEE72 pKa = 4.09AAKK75 pKa = 10.52NRR77 pKa = 11.84DD78 pKa = 2.96WQEE81 pKa = 3.78YY82 pKa = 7.88MSNTAHH88 pKa = 5.58QRR90 pKa = 11.84EE91 pKa = 4.28VRR93 pKa = 11.84DD94 pKa = 3.79LKK96 pKa = 11.22AAGLNPILSAMGGNGAAVGSGATASGVTSAGAKK129 pKa = 9.73GDD131 pKa = 3.66RR132 pKa = 11.84DD133 pKa = 3.56TSANNAIVSVLGSLLNSMTSMAMSTNSAITNLAVADD169 pKa = 4.64KK170 pKa = 9.78YY171 pKa = 11.7NAMSKK176 pKa = 8.76YY177 pKa = 9.08TADD180 pKa = 3.77LSSNTQLAGYY190 pKa = 9.18RR191 pKa = 11.84ISAEE195 pKa = 4.14TALNTANINAAVSRR209 pKa = 11.84YY210 pKa = 9.99VSDD213 pKa = 4.06NNLKK217 pKa = 10.54GSQAMAAATKK227 pKa = 10.37ISAQLHH233 pKa = 6.0ADD235 pKa = 3.32ASKK238 pKa = 9.72YY239 pKa = 10.67AADD242 pKa = 3.88KK243 pKa = 11.21GYY245 pKa = 8.79LTSTDD250 pKa = 3.3VAKK253 pKa = 10.81INADD257 pKa = 2.93INKK260 pKa = 8.89QLKK263 pKa = 9.76QMGIDD268 pKa = 3.42AQFDD272 pKa = 3.6FARR275 pKa = 11.84DD276 pKa = 3.64YY277 pKa = 11.14PSNAFQAVGSISDD290 pKa = 3.74LFGNASGGSGKK301 pKa = 10.5GVSSLFNSVVNGLRR315 pKa = 11.84GGFGEE320 pKa = 4.41RR321 pKa = 11.84KK322 pKa = 9.66SGFSGGSKK330 pKa = 10.15GYY332 pKa = 9.6SGKK335 pKa = 10.44RR336 pKa = 3.21
Molecular weight: 34.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1246 |
81 |
558 |
311.5 |
34.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.433 ± 2.352 | 0.401 ± 0.341 |
6.982 ± 0.434 | 4.173 ± 1.049 |
3.772 ± 0.606 | 7.223 ± 0.887 |
1.605 ± 0.266 | 4.735 ± 0.474 |
5.217 ± 1.038 | 6.742 ± 0.756 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.809 ± 0.537 | 5.136 ± 1.307 |
5.056 ± 1.438 | 3.692 ± 0.51 |
4.815 ± 0.479 | 8.668 ± 1.338 |
6.18 ± 0.556 | 6.019 ± 1.122 |
1.525 ± 0.336 | 4.815 ± 0.933 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
