
Mycolicibacterium holsaticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
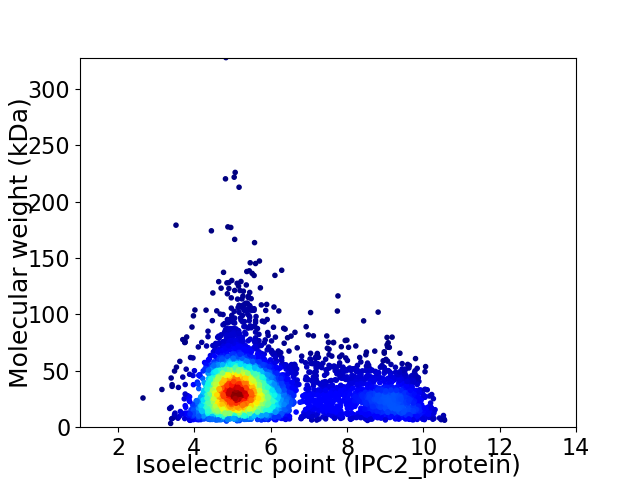
Virtual 2D-PAGE plot for 5158 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
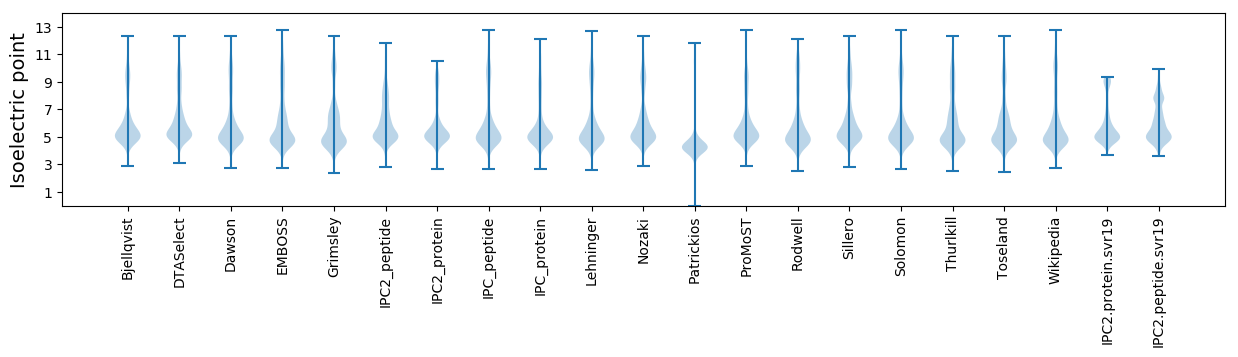
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3S0X4|A0A1E3S0X4_9MYCO GTPase Obg OS=Mycolicibacterium holsaticum OX=152142 GN=obg PE=3 SV=1
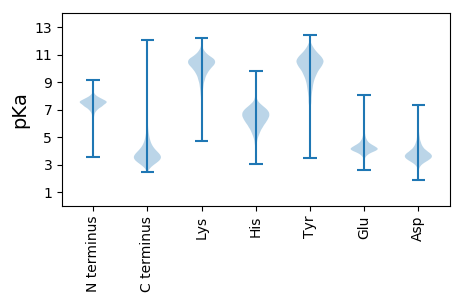
MM1 pKa = 7.21NRR3 pKa = 11.84PYY5 pKa = 9.9TAQSPYY11 pKa = 11.09SLTPTEE17 pKa = 4.33RR18 pKa = 11.84FGTYY22 pKa = 9.98AGEE25 pKa = 4.26PPVDD29 pKa = 3.62EE30 pKa = 5.66LDD32 pKa = 4.37YY33 pKa = 11.22PDD35 pKa = 5.86DD36 pKa = 4.3DD37 pKa = 4.54AGYY40 pKa = 10.05RR41 pKa = 11.84SSYY44 pKa = 9.09DD45 pKa = 2.96TGYY48 pKa = 9.03DD49 pKa = 2.79TGYY52 pKa = 11.05DD53 pKa = 3.34ADD55 pKa = 4.49YY56 pKa = 11.33DD57 pKa = 3.95DD58 pKa = 5.53TYY60 pKa = 11.86DD61 pKa = 3.76EE62 pKa = 4.75YY63 pKa = 11.79ADD65 pKa = 4.18EE66 pKa = 4.5YY67 pKa = 11.57ADD69 pKa = 5.02FEE71 pKa = 5.35DD72 pKa = 5.2DD73 pKa = 3.01EE74 pKa = 5.05ARR76 pKa = 11.84IDD78 pKa = 4.27RR79 pKa = 11.84RR80 pKa = 11.84WMWVAGIAGIILFIAVGTTGVILGGGDD107 pKa = 3.48SGSVTATGTSTAPSPSATATSAPRR131 pKa = 11.84ATAPAAPLVPSLPPEE146 pKa = 4.16TVTTVTPTAEE156 pKa = 4.29VTPTAQPVVPPAQAAPPPPAPSPRR180 pKa = 11.84TVTYY184 pKa = 10.07RR185 pKa = 11.84VTGNRR190 pKa = 11.84QLIDD194 pKa = 4.22LVTVIYY200 pKa = 10.54TDD202 pKa = 3.03AQGALRR208 pKa = 11.84TDD210 pKa = 3.67VNVALPWAKK219 pKa = 9.24TVVLDD224 pKa = 4.29PGVEE228 pKa = 4.03LSSVTATSVAGQLNCSITDD247 pKa = 3.3ASGALIAAQNNNTIITNCTKK267 pKa = 10.95
MM1 pKa = 7.21NRR3 pKa = 11.84PYY5 pKa = 9.9TAQSPYY11 pKa = 11.09SLTPTEE17 pKa = 4.33RR18 pKa = 11.84FGTYY22 pKa = 9.98AGEE25 pKa = 4.26PPVDD29 pKa = 3.62EE30 pKa = 5.66LDD32 pKa = 4.37YY33 pKa = 11.22PDD35 pKa = 5.86DD36 pKa = 4.3DD37 pKa = 4.54AGYY40 pKa = 10.05RR41 pKa = 11.84SSYY44 pKa = 9.09DD45 pKa = 2.96TGYY48 pKa = 9.03DD49 pKa = 2.79TGYY52 pKa = 11.05DD53 pKa = 3.34ADD55 pKa = 4.49YY56 pKa = 11.33DD57 pKa = 3.95DD58 pKa = 5.53TYY60 pKa = 11.86DD61 pKa = 3.76EE62 pKa = 4.75YY63 pKa = 11.79ADD65 pKa = 4.18EE66 pKa = 4.5YY67 pKa = 11.57ADD69 pKa = 5.02FEE71 pKa = 5.35DD72 pKa = 5.2DD73 pKa = 3.01EE74 pKa = 5.05ARR76 pKa = 11.84IDD78 pKa = 4.27RR79 pKa = 11.84RR80 pKa = 11.84WMWVAGIAGIILFIAVGTTGVILGGGDD107 pKa = 3.48SGSVTATGTSTAPSPSATATSAPRR131 pKa = 11.84ATAPAAPLVPSLPPEE146 pKa = 4.16TVTTVTPTAEE156 pKa = 4.29VTPTAQPVVPPAQAAPPPPAPSPRR180 pKa = 11.84TVTYY184 pKa = 10.07RR185 pKa = 11.84VTGNRR190 pKa = 11.84QLIDD194 pKa = 4.22LVTVIYY200 pKa = 10.54TDD202 pKa = 3.03AQGALRR208 pKa = 11.84TDD210 pKa = 3.67VNVALPWAKK219 pKa = 9.24TVVLDD224 pKa = 4.29PGVEE228 pKa = 4.03LSSVTATSVAGQLNCSITDD247 pKa = 3.3ASGALIAAQNNNTIITNCTKK267 pKa = 10.95
Molecular weight: 27.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3S0U2|A0A1E3S0U2_9MYCO TetR family transcriptional regulator OS=Mycolicibacterium holsaticum OX=152142 GN=BHQ17_05660 PE=4 SV=1
MM1 pKa = 7.61AHH3 pKa = 6.91RR4 pKa = 11.84KK5 pKa = 9.24KK6 pKa = 10.93GPSRR10 pKa = 11.84RR11 pKa = 11.84SGLSGVAVPRR21 pKa = 11.84AQARR25 pKa = 11.84ALHH28 pKa = 6.24SVDD31 pKa = 2.82IHH33 pKa = 6.64PPSSTDD39 pKa = 2.76GSFWGRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84VLLLNSTYY55 pKa = 10.82EE56 pKa = 4.11PLTALPMRR64 pKa = 11.84RR65 pKa = 11.84AVIMLMCGKK74 pKa = 10.39ADD76 pKa = 3.66VVHH79 pKa = 7.73DD80 pKa = 4.4DD81 pKa = 3.46PGGPVIHH88 pKa = 6.67SATRR92 pKa = 11.84TIVVPSVIRR101 pKa = 11.84LKK103 pKa = 10.81VYY105 pKa = 10.18VRR107 pKa = 11.84VPYY110 pKa = 10.07RR111 pKa = 11.84ARR113 pKa = 11.84VPMTRR118 pKa = 11.84AALMHH123 pKa = 6.99RR124 pKa = 11.84DD125 pKa = 3.56RR126 pKa = 11.84FRR128 pKa = 11.84CAYY131 pKa = 10.03CGGKK135 pKa = 10.0ADD137 pKa = 4.1TVDD140 pKa = 3.3HH141 pKa = 5.56VVPRR145 pKa = 11.84SRR147 pKa = 11.84GGEE150 pKa = 3.98HH151 pKa = 5.99SWEE154 pKa = 4.09NCVAACSTCNHH165 pKa = 6.31RR166 pKa = 11.84KK167 pKa = 9.52ADD169 pKa = 4.13RR170 pKa = 11.84LLGEE174 pKa = 4.82LGWTLRR180 pKa = 11.84STPLPPKK187 pKa = 8.18GQHH190 pKa = 4.74WRR192 pKa = 11.84LLSTVKK198 pKa = 10.49EE199 pKa = 4.39LDD201 pKa = 4.0PSWMRR206 pKa = 11.84YY207 pKa = 9.3LGEE210 pKa = 4.2GAAA213 pKa = 4.63
MM1 pKa = 7.61AHH3 pKa = 6.91RR4 pKa = 11.84KK5 pKa = 9.24KK6 pKa = 10.93GPSRR10 pKa = 11.84RR11 pKa = 11.84SGLSGVAVPRR21 pKa = 11.84AQARR25 pKa = 11.84ALHH28 pKa = 6.24SVDD31 pKa = 2.82IHH33 pKa = 6.64PPSSTDD39 pKa = 2.76GSFWGRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84VLLLNSTYY55 pKa = 10.82EE56 pKa = 4.11PLTALPMRR64 pKa = 11.84RR65 pKa = 11.84AVIMLMCGKK74 pKa = 10.39ADD76 pKa = 3.66VVHH79 pKa = 7.73DD80 pKa = 4.4DD81 pKa = 3.46PGGPVIHH88 pKa = 6.67SATRR92 pKa = 11.84TIVVPSVIRR101 pKa = 11.84LKK103 pKa = 10.81VYY105 pKa = 10.18VRR107 pKa = 11.84VPYY110 pKa = 10.07RR111 pKa = 11.84ARR113 pKa = 11.84VPMTRR118 pKa = 11.84AALMHH123 pKa = 6.99RR124 pKa = 11.84DD125 pKa = 3.56RR126 pKa = 11.84FRR128 pKa = 11.84CAYY131 pKa = 10.03CGGKK135 pKa = 10.0ADD137 pKa = 4.1TVDD140 pKa = 3.3HH141 pKa = 5.56VVPRR145 pKa = 11.84SRR147 pKa = 11.84GGEE150 pKa = 3.98HH151 pKa = 5.99SWEE154 pKa = 4.09NCVAACSTCNHH165 pKa = 6.31RR166 pKa = 11.84KK167 pKa = 9.52ADD169 pKa = 4.13RR170 pKa = 11.84LLGEE174 pKa = 4.82LGWTLRR180 pKa = 11.84STPLPPKK187 pKa = 8.18GQHH190 pKa = 4.74WRR192 pKa = 11.84LLSTVKK198 pKa = 10.49EE199 pKa = 4.39LDD201 pKa = 4.0PSWMRR206 pKa = 11.84YY207 pKa = 9.3LGEE210 pKa = 4.2GAAA213 pKa = 4.63
Molecular weight: 23.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1625613 |
30 |
3079 |
315.2 |
33.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.855 ± 0.045 | 0.84 ± 0.01 |
6.444 ± 0.032 | 5.335 ± 0.031 |
3.098 ± 0.019 | 8.715 ± 0.035 |
2.177 ± 0.017 | 4.253 ± 0.019 |
2.233 ± 0.02 | 9.691 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.126 ± 0.012 | 2.296 ± 0.018 |
5.843 ± 0.034 | 3.084 ± 0.02 |
7.237 ± 0.035 | 5.235 ± 0.018 |
6.056 ± 0.022 | 8.771 ± 0.035 |
1.509 ± 0.014 | 2.201 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
