
Dragonfly associated cyclovirus 8
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
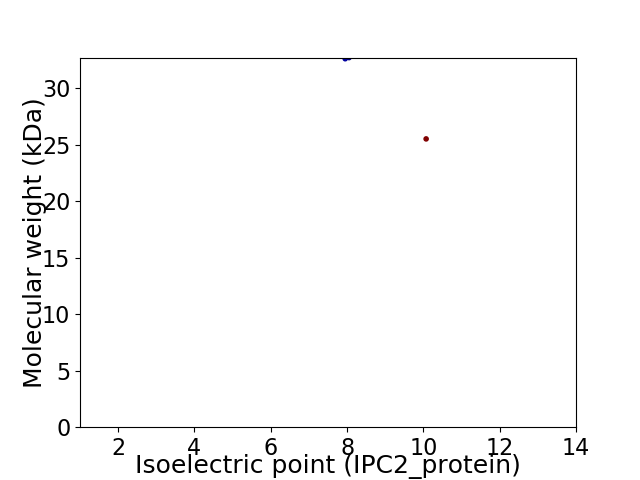
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
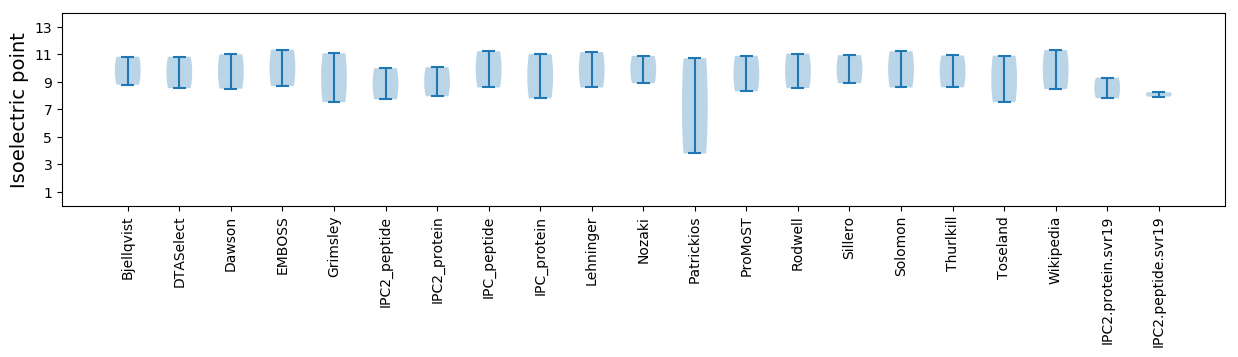
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9V9N2|M9V9N2_9CIRC ATP-dependent helicase Rep OS=Dragonfly associated cyclovirus 8 OX=1574362 PE=3 SV=1
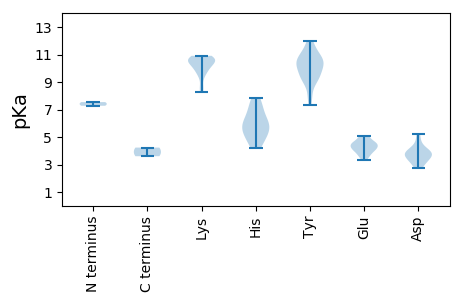
MM1 pKa = 7.54CNSTVRR7 pKa = 11.84KK8 pKa = 9.52FCFTWNNYY16 pKa = 7.32TEE18 pKa = 4.62HH19 pKa = 7.81DD20 pKa = 3.93EE21 pKa = 4.44NKK23 pKa = 10.2CKK25 pKa = 10.84DD26 pKa = 4.81FIAQYY31 pKa = 10.03CKK33 pKa = 10.59YY34 pKa = 10.45GIFGKK39 pKa = 10.35EE40 pKa = 3.71LAPTTNTPHH49 pKa = 6.25LQGYY53 pKa = 9.2CNLSKK58 pKa = 10.65PMRR61 pKa = 11.84FSTIKK66 pKa = 10.19KK67 pKa = 8.25HH68 pKa = 5.12LHH70 pKa = 5.16NSIHH74 pKa = 6.15IEE76 pKa = 3.86KK77 pKa = 10.82ANGSDD82 pKa = 3.63EE83 pKa = 4.12QNKK86 pKa = 9.6EE87 pKa = 4.02YY88 pKa = 10.39CSKK91 pKa = 10.72SGEE94 pKa = 4.12IFEE97 pKa = 5.06KK98 pKa = 9.94GTPIKK103 pKa = 10.38RR104 pKa = 11.84GQRR107 pKa = 11.84TDD109 pKa = 3.62LQSLLADD116 pKa = 3.37IQDD119 pKa = 3.91GNRR122 pKa = 11.84NIQTLAQSHH131 pKa = 5.76PTTYY135 pKa = 9.72IRR137 pKa = 11.84YY138 pKa = 8.25FRR140 pKa = 11.84GIHH143 pKa = 5.69TYY145 pKa = 11.14LNLVHH150 pKa = 7.2PIAPRR155 pKa = 11.84NFKK158 pKa = 10.11TDD160 pKa = 2.71TYY162 pKa = 10.15YY163 pKa = 11.14YY164 pKa = 9.18WGPPGSGKK172 pKa = 8.76SRR174 pKa = 11.84RR175 pKa = 11.84ALEE178 pKa = 4.02EE179 pKa = 3.36ATARR183 pKa = 11.84CNEE186 pKa = 4.13SIYY189 pKa = 10.53YY190 pKa = 9.62KK191 pKa = 10.54PRR193 pKa = 11.84GQWWDD198 pKa = 3.68GYY200 pKa = 9.12HH201 pKa = 4.78QQEE204 pKa = 4.6GVIIDD209 pKa = 5.22DD210 pKa = 4.3FYY212 pKa = 11.96GWIKK216 pKa = 10.71YY217 pKa = 10.27DD218 pKa = 3.78EE219 pKa = 4.42LLKK222 pKa = 10.49VTDD225 pKa = 4.36RR226 pKa = 11.84YY227 pKa = 9.86PYY229 pKa = 9.33KK230 pKa = 10.68VQVKK234 pKa = 10.33GSFEE238 pKa = 4.21EE239 pKa = 4.8FTSKK243 pKa = 10.73HH244 pKa = 4.18IWITSNVDD252 pKa = 2.75TCDD255 pKa = 3.11LYY257 pKa = 11.61KK258 pKa = 10.91FIGYY262 pKa = 7.93CTDD265 pKa = 4.06AIEE268 pKa = 5.0RR269 pKa = 11.84RR270 pKa = 11.84ITLKK274 pKa = 10.85SYY276 pKa = 10.13MSS278 pKa = 3.6
MM1 pKa = 7.54CNSTVRR7 pKa = 11.84KK8 pKa = 9.52FCFTWNNYY16 pKa = 7.32TEE18 pKa = 4.62HH19 pKa = 7.81DD20 pKa = 3.93EE21 pKa = 4.44NKK23 pKa = 10.2CKK25 pKa = 10.84DD26 pKa = 4.81FIAQYY31 pKa = 10.03CKK33 pKa = 10.59YY34 pKa = 10.45GIFGKK39 pKa = 10.35EE40 pKa = 3.71LAPTTNTPHH49 pKa = 6.25LQGYY53 pKa = 9.2CNLSKK58 pKa = 10.65PMRR61 pKa = 11.84FSTIKK66 pKa = 10.19KK67 pKa = 8.25HH68 pKa = 5.12LHH70 pKa = 5.16NSIHH74 pKa = 6.15IEE76 pKa = 3.86KK77 pKa = 10.82ANGSDD82 pKa = 3.63EE83 pKa = 4.12QNKK86 pKa = 9.6EE87 pKa = 4.02YY88 pKa = 10.39CSKK91 pKa = 10.72SGEE94 pKa = 4.12IFEE97 pKa = 5.06KK98 pKa = 9.94GTPIKK103 pKa = 10.38RR104 pKa = 11.84GQRR107 pKa = 11.84TDD109 pKa = 3.62LQSLLADD116 pKa = 3.37IQDD119 pKa = 3.91GNRR122 pKa = 11.84NIQTLAQSHH131 pKa = 5.76PTTYY135 pKa = 9.72IRR137 pKa = 11.84YY138 pKa = 8.25FRR140 pKa = 11.84GIHH143 pKa = 5.69TYY145 pKa = 11.14LNLVHH150 pKa = 7.2PIAPRR155 pKa = 11.84NFKK158 pKa = 10.11TDD160 pKa = 2.71TYY162 pKa = 10.15YY163 pKa = 11.14YY164 pKa = 9.18WGPPGSGKK172 pKa = 8.76SRR174 pKa = 11.84RR175 pKa = 11.84ALEE178 pKa = 4.02EE179 pKa = 3.36ATARR183 pKa = 11.84CNEE186 pKa = 4.13SIYY189 pKa = 10.53YY190 pKa = 9.62KK191 pKa = 10.54PRR193 pKa = 11.84GQWWDD198 pKa = 3.68GYY200 pKa = 9.12HH201 pKa = 4.78QQEE204 pKa = 4.6GVIIDD209 pKa = 5.22DD210 pKa = 4.3FYY212 pKa = 11.96GWIKK216 pKa = 10.71YY217 pKa = 10.27DD218 pKa = 3.78EE219 pKa = 4.42LLKK222 pKa = 10.49VTDD225 pKa = 4.36RR226 pKa = 11.84YY227 pKa = 9.86PYY229 pKa = 9.33KK230 pKa = 10.68VQVKK234 pKa = 10.33GSFEE238 pKa = 4.21EE239 pKa = 4.8FTSKK243 pKa = 10.73HH244 pKa = 4.18IWITSNVDD252 pKa = 2.75TCDD255 pKa = 3.11LYY257 pKa = 11.61KK258 pKa = 10.91FIGYY262 pKa = 7.93CTDD265 pKa = 4.06AIEE268 pKa = 5.0RR269 pKa = 11.84RR270 pKa = 11.84ITLKK274 pKa = 10.85SYY276 pKa = 10.13MSS278 pKa = 3.6
Molecular weight: 32.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9V9N2|M9V9N2_9CIRC ATP-dependent helicase Rep OS=Dragonfly associated cyclovirus 8 OX=1574362 PE=3 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84YY4 pKa = 9.13RR5 pKa = 11.84RR6 pKa = 11.84LRR8 pKa = 11.84RR9 pKa = 11.84PLLRR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84SSRR19 pKa = 11.84LRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 8.47PLRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84FRR31 pKa = 11.84FKK33 pKa = 9.86RR34 pKa = 11.84TPVLYY39 pKa = 10.66VKK41 pKa = 9.37LTRR44 pKa = 11.84TVQVVSVINKK54 pKa = 9.88GGLNSLNQVLNDD66 pKa = 3.43FAEE69 pKa = 4.58FQNLAPSFEE78 pKa = 4.19RR79 pKa = 11.84VKK81 pKa = 10.79VYY83 pKa = 10.65RR84 pKa = 11.84LNVRR88 pKa = 11.84VFPHH92 pKa = 5.84QNVSNNSTSKK102 pKa = 9.83VPGYY106 pKa = 10.88AIVPYY111 pKa = 10.07HH112 pKa = 6.98RR113 pKa = 11.84PPPAATPSFSACLSIDD129 pKa = 3.19RR130 pKa = 11.84AKK132 pKa = 10.65VYY134 pKa = 10.52RR135 pKa = 11.84GTAFGRR141 pKa = 11.84MSLIPASRR149 pKa = 11.84LDD151 pKa = 3.52VEE153 pKa = 5.1ANTGEE158 pKa = 4.53KK159 pKa = 10.42YY160 pKa = 10.8SPDD163 pKa = 3.92RR164 pKa = 11.84IDD166 pKa = 3.21WKK168 pKa = 10.79PEE170 pKa = 3.65FEE172 pKa = 4.36ISSAAATQYY181 pKa = 11.02LYY183 pKa = 10.54TGFVIFEE190 pKa = 4.44TEE192 pKa = 3.58PTAPDD197 pKa = 3.97SEE199 pKa = 4.57TATYY203 pKa = 9.14TIVYY207 pKa = 9.6DD208 pKa = 3.79LYY210 pKa = 11.76VRR212 pKa = 11.84FKK214 pKa = 10.4NQRR217 pKa = 11.84SFII220 pKa = 4.23
MM1 pKa = 7.27ARR3 pKa = 11.84YY4 pKa = 9.13RR5 pKa = 11.84RR6 pKa = 11.84LRR8 pKa = 11.84RR9 pKa = 11.84PLLRR13 pKa = 11.84RR14 pKa = 11.84NRR16 pKa = 11.84SSRR19 pKa = 11.84LRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 8.47PLRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84FRR31 pKa = 11.84FKK33 pKa = 9.86RR34 pKa = 11.84TPVLYY39 pKa = 10.66VKK41 pKa = 9.37LTRR44 pKa = 11.84TVQVVSVINKK54 pKa = 9.88GGLNSLNQVLNDD66 pKa = 3.43FAEE69 pKa = 4.58FQNLAPSFEE78 pKa = 4.19RR79 pKa = 11.84VKK81 pKa = 10.79VYY83 pKa = 10.65RR84 pKa = 11.84LNVRR88 pKa = 11.84VFPHH92 pKa = 5.84QNVSNNSTSKK102 pKa = 9.83VPGYY106 pKa = 10.88AIVPYY111 pKa = 10.07HH112 pKa = 6.98RR113 pKa = 11.84PPPAATPSFSACLSIDD129 pKa = 3.19RR130 pKa = 11.84AKK132 pKa = 10.65VYY134 pKa = 10.52RR135 pKa = 11.84GTAFGRR141 pKa = 11.84MSLIPASRR149 pKa = 11.84LDD151 pKa = 3.52VEE153 pKa = 5.1ANTGEE158 pKa = 4.53KK159 pKa = 10.42YY160 pKa = 10.8SPDD163 pKa = 3.92RR164 pKa = 11.84IDD166 pKa = 3.21WKK168 pKa = 10.79PEE170 pKa = 3.65FEE172 pKa = 4.36ISSAAATQYY181 pKa = 11.02LYY183 pKa = 10.54TGFVIFEE190 pKa = 4.44TEE192 pKa = 3.58PTAPDD197 pKa = 3.97SEE199 pKa = 4.57TATYY203 pKa = 9.14TIVYY207 pKa = 9.6DD208 pKa = 3.79LYY210 pKa = 11.76VRR212 pKa = 11.84FKK214 pKa = 10.4NQRR217 pKa = 11.84SFII220 pKa = 4.23
Molecular weight: 25.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
498 |
220 |
278 |
249.0 |
29.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.221 ± 1.32 | 2.008 ± 0.999 |
4.418 ± 0.795 | 5.02 ± 0.598 |
5.02 ± 0.572 | 5.02 ± 1.183 |
2.41 ± 0.965 | 6.024 ± 1.244 |
6.627 ± 1.339 | 6.426 ± 0.837 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.004 ± 0.061 | 5.422 ± 0.021 |
5.422 ± 1.191 | 3.614 ± 0.571 |
8.835 ± 2.796 | 7.028 ± 0.742 |
7.229 ± 0.557 | 5.221 ± 2.197 |
1.406 ± 0.612 | 6.627 ± 0.754 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
