
Beihai barnacle virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.44
Get precalculated fractions of proteins
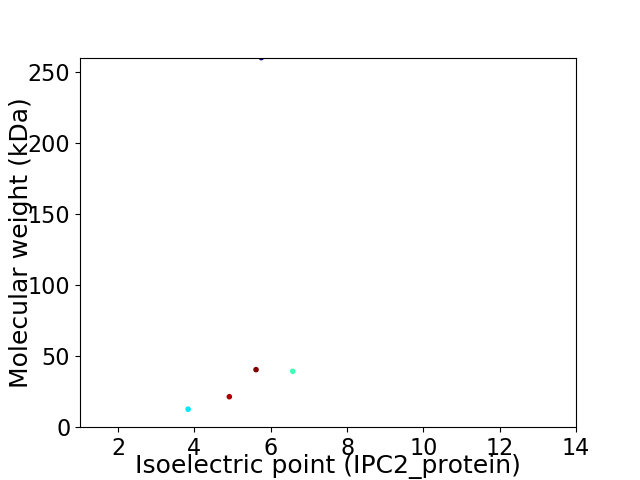
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJS1|A0A1L3KJS1_9VIRU Uncharacterized protein OS=Beihai barnacle virus 3 OX=1922361 PE=4 SV=1
MM1 pKa = 7.14FRR3 pKa = 11.84ASLLQVSPPLYY14 pKa = 8.88EE15 pKa = 4.0WPVADD20 pKa = 4.81AEE22 pKa = 4.59VRR24 pKa = 11.84DD25 pKa = 4.1STASIDD31 pKa = 3.58SGINQPVLGVKK42 pKa = 10.41AILGKK47 pKa = 10.41IKK49 pKa = 10.68DD50 pKa = 3.88LTEE53 pKa = 4.81LIEE56 pKa = 4.49GEE58 pKa = 4.26IVDD61 pKa = 4.32LAVVVDD67 pKa = 4.81SIEE70 pKa = 4.03ATTLEE75 pKa = 4.13IAASVDD81 pKa = 3.82VIAGEE86 pKa = 3.89ITEE89 pKa = 4.25IAIATTDD96 pKa = 3.65CAVALDD102 pKa = 4.68SIAVTVDD109 pKa = 2.74LHH111 pKa = 7.01NVVPWPCVGII121 pKa = 4.13
MM1 pKa = 7.14FRR3 pKa = 11.84ASLLQVSPPLYY14 pKa = 8.88EE15 pKa = 4.0WPVADD20 pKa = 4.81AEE22 pKa = 4.59VRR24 pKa = 11.84DD25 pKa = 4.1STASIDD31 pKa = 3.58SGINQPVLGVKK42 pKa = 10.41AILGKK47 pKa = 10.41IKK49 pKa = 10.68DD50 pKa = 3.88LTEE53 pKa = 4.81LIEE56 pKa = 4.49GEE58 pKa = 4.26IVDD61 pKa = 4.32LAVVVDD67 pKa = 4.81SIEE70 pKa = 4.03ATTLEE75 pKa = 4.13IAASVDD81 pKa = 3.82VIAGEE86 pKa = 3.89ITEE89 pKa = 4.25IAIATTDD96 pKa = 3.65CAVALDD102 pKa = 4.68SIAVTVDD109 pKa = 2.74LHH111 pKa = 7.01NVVPWPCVGII121 pKa = 4.13
Molecular weight: 12.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJJ2|A0A1L3KJJ2_9VIRU Uncharacterized protein OS=Beihai barnacle virus 3 OX=1922361 PE=4 SV=1
MM1 pKa = 7.52PRR3 pKa = 11.84FGRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FAPRR12 pKa = 11.84RR13 pKa = 11.84GRR15 pKa = 11.84ARR17 pKa = 11.84GGPARR22 pKa = 11.84PQNVTVNIKK31 pKa = 10.56LSQTDD36 pKa = 3.46ANADD40 pKa = 3.45GSNPAVYY47 pKa = 10.39VSLSSQQIIDD57 pKa = 4.06HH58 pKa = 6.34GPVLNGIKK66 pKa = 9.89PAFARR71 pKa = 11.84IRR73 pKa = 11.84GMTLEE78 pKa = 4.35VTSGAAATTEE88 pKa = 4.49GIGAYY93 pKa = 7.3MAWRR97 pKa = 11.84GVMPDD102 pKa = 3.58TTTMDD107 pKa = 3.68LGATILEE114 pKa = 4.41AATRR118 pKa = 11.84RR119 pKa = 11.84SAPGISARR127 pKa = 11.84HH128 pKa = 4.61TWRR131 pKa = 11.84NFPTRR136 pKa = 11.84IRR138 pKa = 11.84LRR140 pKa = 11.84TQGAEE145 pKa = 3.82RR146 pKa = 11.84TDD148 pKa = 3.12IQGNVSHH155 pKa = 7.22HH156 pKa = 6.94DD157 pKa = 3.53VFGVGVTYY165 pKa = 10.03PKK167 pKa = 10.56GAPPPLAILKK177 pKa = 8.67ATFQYY182 pKa = 10.9GGADD186 pKa = 3.17MVSAEE191 pKa = 4.26VVEE194 pKa = 4.53GTGEE198 pKa = 3.58IEE200 pKa = 4.09YY201 pKa = 10.48DD202 pKa = 3.19HH203 pKa = 7.2EE204 pKa = 4.46YY205 pKa = 10.03PAEE208 pKa = 4.1KK209 pKa = 9.35LTRR212 pKa = 11.84DD213 pKa = 3.84FLDD216 pKa = 3.38QANEE220 pKa = 3.92KK221 pKa = 10.7LGIPLGYY228 pKa = 9.2TSDD231 pKa = 3.35SGAICFVRR239 pKa = 11.84MLVVYY244 pKa = 10.68LCDD247 pKa = 3.68SKK249 pKa = 11.36EE250 pKa = 3.97LYY252 pKa = 10.23AVNSGALTSFKK263 pKa = 10.67LPYY266 pKa = 9.99EE267 pKa = 4.24SFVFPARR274 pKa = 11.84SSSNEE279 pKa = 3.14GKK281 pKa = 10.5LYY283 pKa = 10.82SRR285 pKa = 11.84INAWCQTPITPPGAAPGEE303 pKa = 4.62VYY305 pKa = 9.68PAHH308 pKa = 7.22ADD310 pKa = 3.62DD311 pKa = 4.4VNLVGDD317 pKa = 4.5FEE319 pKa = 6.15HH320 pKa = 7.69DD321 pKa = 3.73FGIHH325 pKa = 6.32DD326 pKa = 3.83MYY328 pKa = 10.95VRR330 pKa = 11.84SVMITPMTRR339 pKa = 11.84TAGEE343 pKa = 4.52VLTMAASGAAAVATYY358 pKa = 9.2PVIPASSSS366 pKa = 3.02
MM1 pKa = 7.52PRR3 pKa = 11.84FGRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FAPRR12 pKa = 11.84RR13 pKa = 11.84GRR15 pKa = 11.84ARR17 pKa = 11.84GGPARR22 pKa = 11.84PQNVTVNIKK31 pKa = 10.56LSQTDD36 pKa = 3.46ANADD40 pKa = 3.45GSNPAVYY47 pKa = 10.39VSLSSQQIIDD57 pKa = 4.06HH58 pKa = 6.34GPVLNGIKK66 pKa = 9.89PAFARR71 pKa = 11.84IRR73 pKa = 11.84GMTLEE78 pKa = 4.35VTSGAAATTEE88 pKa = 4.49GIGAYY93 pKa = 7.3MAWRR97 pKa = 11.84GVMPDD102 pKa = 3.58TTTMDD107 pKa = 3.68LGATILEE114 pKa = 4.41AATRR118 pKa = 11.84RR119 pKa = 11.84SAPGISARR127 pKa = 11.84HH128 pKa = 4.61TWRR131 pKa = 11.84NFPTRR136 pKa = 11.84IRR138 pKa = 11.84LRR140 pKa = 11.84TQGAEE145 pKa = 3.82RR146 pKa = 11.84TDD148 pKa = 3.12IQGNVSHH155 pKa = 7.22HH156 pKa = 6.94DD157 pKa = 3.53VFGVGVTYY165 pKa = 10.03PKK167 pKa = 10.56GAPPPLAILKK177 pKa = 8.67ATFQYY182 pKa = 10.9GGADD186 pKa = 3.17MVSAEE191 pKa = 4.26VVEE194 pKa = 4.53GTGEE198 pKa = 3.58IEE200 pKa = 4.09YY201 pKa = 10.48DD202 pKa = 3.19HH203 pKa = 7.2EE204 pKa = 4.46YY205 pKa = 10.03PAEE208 pKa = 4.1KK209 pKa = 9.35LTRR212 pKa = 11.84DD213 pKa = 3.84FLDD216 pKa = 3.38QANEE220 pKa = 3.92KK221 pKa = 10.7LGIPLGYY228 pKa = 9.2TSDD231 pKa = 3.35SGAICFVRR239 pKa = 11.84MLVVYY244 pKa = 10.68LCDD247 pKa = 3.68SKK249 pKa = 11.36EE250 pKa = 3.97LYY252 pKa = 10.23AVNSGALTSFKK263 pKa = 10.67LPYY266 pKa = 9.99EE267 pKa = 4.24SFVFPARR274 pKa = 11.84SSSNEE279 pKa = 3.14GKK281 pKa = 10.5LYY283 pKa = 10.82SRR285 pKa = 11.84INAWCQTPITPPGAAPGEE303 pKa = 4.62VYY305 pKa = 9.68PAHH308 pKa = 7.22ADD310 pKa = 3.62DD311 pKa = 4.4VNLVGDD317 pKa = 4.5FEE319 pKa = 6.15HH320 pKa = 7.69DD321 pKa = 3.73FGIHH325 pKa = 6.32DD326 pKa = 3.83MYY328 pKa = 10.95VRR330 pKa = 11.84SVMITPMTRR339 pKa = 11.84TAGEE343 pKa = 4.52VLTMAASGAAAVATYY358 pKa = 9.2PVIPASSSS366 pKa = 3.02
Molecular weight: 39.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3376 |
121 |
2315 |
675.2 |
74.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.893 ± 0.733 | 1.718 ± 0.217 |
5.954 ± 0.599 | 6.783 ± 1.27 |
3.347 ± 0.251 | 7.109 ± 0.794 |
2.844 ± 0.274 | 4.976 ± 0.634 |
4.502 ± 0.954 | 7.05 ± 0.739 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.169 ± 0.534 | 3.732 ± 0.367 |
4.769 ± 0.536 | 3.614 ± 0.312 |
6.309 ± 0.618 | 6.309 ± 0.729 |
5.509 ± 0.581 | 7.82 ± 0.802 |
1.422 ± 0.177 | 3.169 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
