
Pseudomonas phage Dobby
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Peduovirinae; Citexvirus; Pseudomonas virus Dobby
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
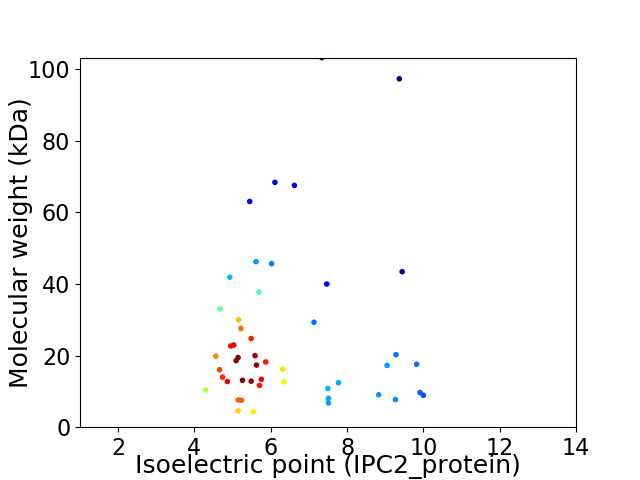
Virtual 2D-PAGE plot for 49 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
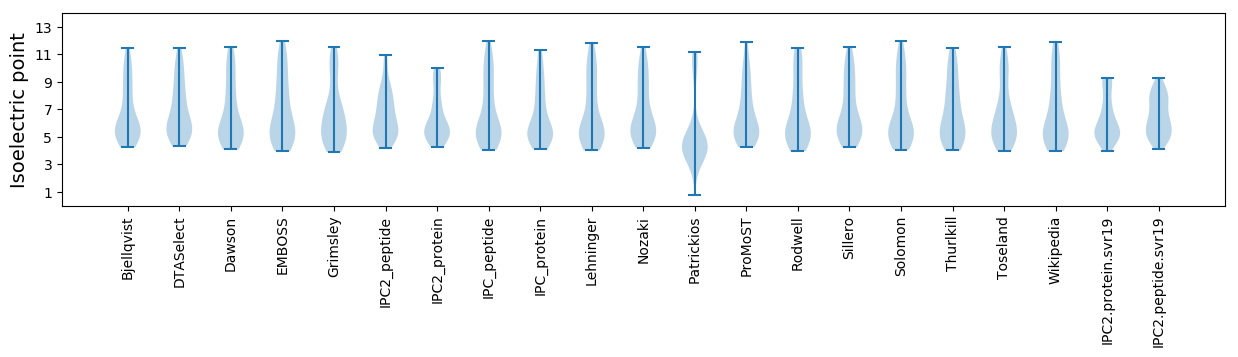
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G8F159|A0A3G8F159_9CAUD Holin family protein OS=Pseudomonas phage Dobby OX=2483611 PE=4 SV=1
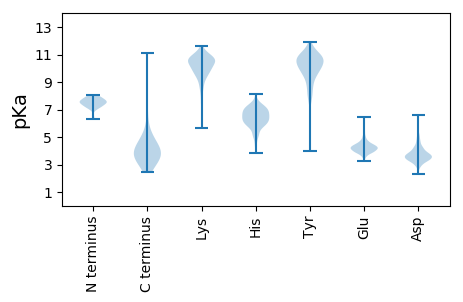
MM1 pKa = 7.23MMLLGFTAIQQATPPTLEE19 pKa = 4.15PLQGIEE25 pKa = 3.95MASNGVIHH33 pKa = 6.71NNYY36 pKa = 8.66TNYY39 pKa = 10.33TYY41 pKa = 11.06DD42 pKa = 3.28FSSSVSASLHH52 pKa = 5.14TDD54 pKa = 2.78EE55 pKa = 5.65HH56 pKa = 7.12LSLEE60 pKa = 4.42AATSAFSSRR69 pKa = 11.84LASMQEE75 pKa = 3.59PLEE78 pKa = 4.08YY79 pKa = 10.05EE80 pKa = 3.99FSKK83 pKa = 10.78ILSDD87 pKa = 4.38NFLDD91 pKa = 4.88LLAA94 pKa = 5.91
MM1 pKa = 7.23MMLLGFTAIQQATPPTLEE19 pKa = 4.15PLQGIEE25 pKa = 3.95MASNGVIHH33 pKa = 6.71NNYY36 pKa = 8.66TNYY39 pKa = 10.33TYY41 pKa = 11.06DD42 pKa = 3.28FSSSVSASLHH52 pKa = 5.14TDD54 pKa = 2.78EE55 pKa = 5.65HH56 pKa = 7.12LSLEE60 pKa = 4.42AATSAFSSRR69 pKa = 11.84LASMQEE75 pKa = 3.59PLEE78 pKa = 4.08YY79 pKa = 10.05EE80 pKa = 3.99FSKK83 pKa = 10.78ILSDD87 pKa = 4.38NFLDD91 pKa = 4.88LLAA94 pKa = 5.91
Molecular weight: 10.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G8F1Q7|A0A3G8F1Q7_9CAUD Tail X protein OS=Pseudomonas phage Dobby OX=2483611 PE=4 SV=1
MM1 pKa = 7.41RR2 pKa = 11.84RR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 9.35RR7 pKa = 11.84KK8 pKa = 8.87HH9 pKa = 6.06NPHH12 pKa = 6.56IPQHH16 pKa = 6.34IDD18 pKa = 2.83QAAIPAAVFFDD29 pKa = 3.54HH30 pKa = 7.22RR31 pKa = 11.84GNGVWYY37 pKa = 8.22TLHH40 pKa = 7.06YY41 pKa = 10.92DD42 pKa = 2.96EE43 pKa = 6.25GGRR46 pKa = 11.84QRR48 pKa = 11.84RR49 pKa = 11.84TNLAPSYY56 pKa = 8.75VTLSEE61 pKa = 3.59LHH63 pKa = 6.75RR64 pKa = 11.84IMEE67 pKa = 4.16EE68 pKa = 3.87RR69 pKa = 11.84EE70 pKa = 4.32GINRR74 pKa = 11.84DD75 pKa = 3.3SLANLCGEE83 pKa = 4.48FHH85 pKa = 7.67KK86 pKa = 10.46STQFKK91 pKa = 10.11RR92 pKa = 11.84LKK94 pKa = 10.37PKK96 pKa = 8.63TQSDD100 pKa = 4.28YY101 pKa = 10.14EE102 pKa = 4.25YY103 pKa = 10.83CRR105 pKa = 11.84EE106 pKa = 3.94VLLAIPTRR114 pKa = 11.84LKK116 pKa = 10.88KK117 pKa = 10.23PLGEE121 pKa = 3.74LAVRR125 pKa = 11.84KK126 pKa = 8.71FSPALVQRR134 pKa = 11.84LVDD137 pKa = 5.17RR138 pKa = 11.84IAEE141 pKa = 4.13EE142 pKa = 4.47GTPSKK147 pKa = 10.33AAHH150 pKa = 5.11VLRR153 pKa = 11.84YY154 pKa = 9.43LRR156 pKa = 11.84RR157 pKa = 11.84VMQWGRR163 pKa = 11.84NRR165 pKa = 11.84GYY167 pKa = 10.87LQINVAQGIEE177 pKa = 4.02APVEE181 pKa = 4.01RR182 pKa = 11.84KK183 pKa = 7.39QRR185 pKa = 11.84RR186 pKa = 11.84LPAPTVMYY194 pKa = 10.54RR195 pKa = 11.84LIDD198 pKa = 3.48RR199 pKa = 11.84ARR201 pKa = 11.84EE202 pKa = 3.8LGKK205 pKa = 10.5LKK207 pKa = 10.41RR208 pKa = 11.84GQPGACPAYY217 pKa = 9.37LCSVMEE223 pKa = 4.12LAYY226 pKa = 10.56LCRR229 pKa = 11.84LRR231 pKa = 11.84GIEE234 pKa = 4.31TVTLSDD240 pKa = 4.09ANEE243 pKa = 4.06LPEE246 pKa = 5.71GILTNRR252 pKa = 11.84RR253 pKa = 11.84KK254 pKa = 10.18GSRR257 pKa = 11.84NNVVRR262 pKa = 11.84WTPRR266 pKa = 11.84LRR268 pKa = 11.84AAWDD272 pKa = 3.13HH273 pKa = 5.77AKK275 pKa = 10.42AYY277 pKa = 7.6RR278 pKa = 11.84TQVWTSKK285 pKa = 10.78SLPTPTAPEE294 pKa = 3.93LRR296 pKa = 11.84PIIVASHH303 pKa = 6.44GGSLQKK309 pKa = 10.77SSLDD313 pKa = 3.71SAWQRR318 pKa = 11.84FITAALEE325 pKa = 4.12AGIITPDD332 pKa = 3.28QRR334 pKa = 11.84FGLHH338 pKa = 5.59DD339 pKa = 4.49LKK341 pKa = 11.09RR342 pKa = 11.84RR343 pKa = 11.84GITDD347 pKa = 3.57TPGTRR352 pKa = 11.84ADD354 pKa = 3.4KK355 pKa = 11.14QEE357 pKa = 4.13ASGHH361 pKa = 5.96RR362 pKa = 11.84DD363 pKa = 2.98EE364 pKa = 6.44SMLDD368 pKa = 3.53VYY370 pKa = 11.02DD371 pKa = 4.53LSVPIVSPSADD382 pKa = 2.92
MM1 pKa = 7.41RR2 pKa = 11.84RR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 9.35RR7 pKa = 11.84KK8 pKa = 8.87HH9 pKa = 6.06NPHH12 pKa = 6.56IPQHH16 pKa = 6.34IDD18 pKa = 2.83QAAIPAAVFFDD29 pKa = 3.54HH30 pKa = 7.22RR31 pKa = 11.84GNGVWYY37 pKa = 8.22TLHH40 pKa = 7.06YY41 pKa = 10.92DD42 pKa = 2.96EE43 pKa = 6.25GGRR46 pKa = 11.84QRR48 pKa = 11.84RR49 pKa = 11.84TNLAPSYY56 pKa = 8.75VTLSEE61 pKa = 3.59LHH63 pKa = 6.75RR64 pKa = 11.84IMEE67 pKa = 4.16EE68 pKa = 3.87RR69 pKa = 11.84EE70 pKa = 4.32GINRR74 pKa = 11.84DD75 pKa = 3.3SLANLCGEE83 pKa = 4.48FHH85 pKa = 7.67KK86 pKa = 10.46STQFKK91 pKa = 10.11RR92 pKa = 11.84LKK94 pKa = 10.37PKK96 pKa = 8.63TQSDD100 pKa = 4.28YY101 pKa = 10.14EE102 pKa = 4.25YY103 pKa = 10.83CRR105 pKa = 11.84EE106 pKa = 3.94VLLAIPTRR114 pKa = 11.84LKK116 pKa = 10.88KK117 pKa = 10.23PLGEE121 pKa = 3.74LAVRR125 pKa = 11.84KK126 pKa = 8.71FSPALVQRR134 pKa = 11.84LVDD137 pKa = 5.17RR138 pKa = 11.84IAEE141 pKa = 4.13EE142 pKa = 4.47GTPSKK147 pKa = 10.33AAHH150 pKa = 5.11VLRR153 pKa = 11.84YY154 pKa = 9.43LRR156 pKa = 11.84RR157 pKa = 11.84VMQWGRR163 pKa = 11.84NRR165 pKa = 11.84GYY167 pKa = 10.87LQINVAQGIEE177 pKa = 4.02APVEE181 pKa = 4.01RR182 pKa = 11.84KK183 pKa = 7.39QRR185 pKa = 11.84RR186 pKa = 11.84LPAPTVMYY194 pKa = 10.54RR195 pKa = 11.84LIDD198 pKa = 3.48RR199 pKa = 11.84ARR201 pKa = 11.84EE202 pKa = 3.8LGKK205 pKa = 10.5LKK207 pKa = 10.41RR208 pKa = 11.84GQPGACPAYY217 pKa = 9.37LCSVMEE223 pKa = 4.12LAYY226 pKa = 10.56LCRR229 pKa = 11.84LRR231 pKa = 11.84GIEE234 pKa = 4.31TVTLSDD240 pKa = 4.09ANEE243 pKa = 4.06LPEE246 pKa = 5.71GILTNRR252 pKa = 11.84RR253 pKa = 11.84KK254 pKa = 10.18GSRR257 pKa = 11.84NNVVRR262 pKa = 11.84WTPRR266 pKa = 11.84LRR268 pKa = 11.84AAWDD272 pKa = 3.13HH273 pKa = 5.77AKK275 pKa = 10.42AYY277 pKa = 7.6RR278 pKa = 11.84TQVWTSKK285 pKa = 10.78SLPTPTAPEE294 pKa = 3.93LRR296 pKa = 11.84PIIVASHH303 pKa = 6.44GGSLQKK309 pKa = 10.77SSLDD313 pKa = 3.71SAWQRR318 pKa = 11.84FITAALEE325 pKa = 4.12AGIITPDD332 pKa = 3.28QRR334 pKa = 11.84FGLHH338 pKa = 5.59DD339 pKa = 4.49LKK341 pKa = 11.09RR342 pKa = 11.84RR343 pKa = 11.84GITDD347 pKa = 3.57TPGTRR352 pKa = 11.84ADD354 pKa = 3.4KK355 pKa = 11.14QEE357 pKa = 4.13ASGHH361 pKa = 5.96RR362 pKa = 11.84DD363 pKa = 2.98EE364 pKa = 6.44SMLDD368 pKa = 3.53VYY370 pKa = 11.02DD371 pKa = 4.53LSVPIVSPSADD382 pKa = 2.92
Molecular weight: 43.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11281 |
38 |
919 |
230.2 |
25.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.338 ± 0.508 | 1.011 ± 0.135 |
5.984 ± 0.235 | 5.664 ± 0.347 |
3.041 ± 0.28 | 7.526 ± 0.423 |
2.003 ± 0.173 | 5.168 ± 0.282 |
4.166 ± 0.335 | 10.673 ± 0.419 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.127 ± 0.185 | 3.227 ± 0.212 |
4.547 ± 0.28 | 4.982 ± 0.29 |
7.021 ± 0.351 | 5.833 ± 0.372 |
5.718 ± 0.299 | 5.611 ± 0.296 |
1.667 ± 0.162 | 2.695 ± 0.145 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
