
Zostera marina amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins
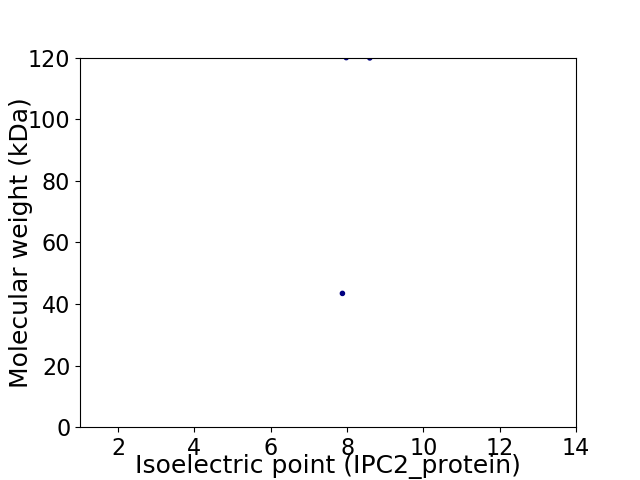
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6R5F0|A0A1W6R5F0_9VIRU Putative coat protein OS=Zostera marina amalgavirus 1 OX=1985202 PE=4 SV=1
MM1 pKa = 7.86AEE3 pKa = 4.71DD4 pKa = 4.62LRR6 pKa = 11.84SQQTLDD12 pKa = 4.41DD13 pKa = 4.27RR14 pKa = 11.84DD15 pKa = 3.9NLKK18 pKa = 10.9LLTDD22 pKa = 4.08AFKK25 pKa = 10.6AYY27 pKa = 9.65PEE29 pKa = 4.95GGLSVVDD36 pKa = 4.12VSLEE40 pKa = 4.15GIAACNYY47 pKa = 6.33TVSRR51 pKa = 11.84AVKK54 pKa = 9.85VMKK57 pKa = 10.04ILKK60 pKa = 9.25PLTKK64 pKa = 10.03NEE66 pKa = 3.86HH67 pKa = 6.22LIKK70 pKa = 10.47LFHH73 pKa = 6.38YY74 pKa = 10.79ANDD77 pKa = 3.25VSNISSILPMTLEE90 pKa = 4.49TNFKK94 pKa = 8.5FCKK97 pKa = 9.62WLTSPVAKK105 pKa = 10.29KK106 pKa = 10.45KK107 pKa = 10.21IAALQNADD115 pKa = 3.56RR116 pKa = 11.84LRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84GSDD123 pKa = 3.08VVTPEE128 pKa = 3.75EE129 pKa = 3.76AAMIALLEE137 pKa = 4.21SAQADD142 pKa = 3.97RR143 pKa = 11.84TTEE146 pKa = 3.86YY147 pKa = 10.44SKK149 pKa = 11.34ARR151 pKa = 11.84ARR153 pKa = 11.84YY154 pKa = 9.35DD155 pKa = 3.14IDD157 pKa = 3.37VAKK160 pKa = 10.4YY161 pKa = 9.17KK162 pKa = 10.79KK163 pKa = 10.3KK164 pKa = 8.77IAKK167 pKa = 7.74RR168 pKa = 11.84TRR170 pKa = 11.84QLEE173 pKa = 3.88EE174 pKa = 5.19DD175 pKa = 3.74LDD177 pKa = 4.27KK178 pKa = 11.69VQASYY183 pKa = 10.83PGLQLIEE190 pKa = 4.79RR191 pKa = 11.84PDD193 pKa = 3.62EE194 pKa = 4.36HH195 pKa = 9.05SVMASAWHH203 pKa = 6.72RR204 pKa = 11.84YY205 pKa = 7.58VDD207 pKa = 4.67FCTSNNFEE215 pKa = 4.29VPQKK219 pKa = 11.16NDD221 pKa = 2.85GNLARR226 pKa = 11.84AYY228 pKa = 9.69KK229 pKa = 9.92QFEE232 pKa = 4.26RR233 pKa = 11.84EE234 pKa = 3.8LTLEE238 pKa = 4.29IKK240 pKa = 9.21NTACQKK246 pKa = 10.61PEE248 pKa = 3.65VRR250 pKa = 11.84DD251 pKa = 3.66YY252 pKa = 11.57LLQYY256 pKa = 10.49CKK258 pKa = 10.72EE259 pKa = 3.98KK260 pKa = 11.17VKK262 pKa = 10.62GFRR265 pKa = 11.84AQFEE269 pKa = 4.55DD270 pKa = 4.96KK271 pKa = 10.56RR272 pKa = 11.84AQTYY276 pKa = 9.35RR277 pKa = 11.84GYY279 pKa = 9.81YY280 pKa = 9.83ACATGDD286 pKa = 3.98ILPEE290 pKa = 3.73DD291 pKa = 4.05TPRR294 pKa = 11.84KK295 pKa = 8.88KK296 pKa = 10.46RR297 pKa = 11.84KK298 pKa = 9.53ASRR301 pKa = 11.84PDD303 pKa = 3.05TGGEE307 pKa = 3.91ANQEE311 pKa = 4.06GEE313 pKa = 4.52DD314 pKa = 3.95DD315 pKa = 4.39SEE317 pKa = 4.94HH318 pKa = 7.56SVIPNLLDD326 pKa = 3.24TCVGDD331 pKa = 6.28DD332 pKa = 3.34RR333 pKa = 11.84RR334 pKa = 11.84AWDD337 pKa = 3.63KK338 pKa = 8.73THH340 pKa = 6.67PTNPEE345 pKa = 3.58KK346 pKa = 10.61PGNDD350 pKa = 3.23SSSDD354 pKa = 3.26EE355 pKa = 4.24SAEE358 pKa = 3.98NQPASTVADD367 pKa = 3.8RR368 pKa = 11.84TRR370 pKa = 11.84TRR372 pKa = 11.84RR373 pKa = 11.84RR374 pKa = 11.84KK375 pKa = 9.68ADD377 pKa = 3.23KK378 pKa = 10.29HH379 pKa = 5.83SKK381 pKa = 9.49KK382 pKa = 10.73
MM1 pKa = 7.86AEE3 pKa = 4.71DD4 pKa = 4.62LRR6 pKa = 11.84SQQTLDD12 pKa = 4.41DD13 pKa = 4.27RR14 pKa = 11.84DD15 pKa = 3.9NLKK18 pKa = 10.9LLTDD22 pKa = 4.08AFKK25 pKa = 10.6AYY27 pKa = 9.65PEE29 pKa = 4.95GGLSVVDD36 pKa = 4.12VSLEE40 pKa = 4.15GIAACNYY47 pKa = 6.33TVSRR51 pKa = 11.84AVKK54 pKa = 9.85VMKK57 pKa = 10.04ILKK60 pKa = 9.25PLTKK64 pKa = 10.03NEE66 pKa = 3.86HH67 pKa = 6.22LIKK70 pKa = 10.47LFHH73 pKa = 6.38YY74 pKa = 10.79ANDD77 pKa = 3.25VSNISSILPMTLEE90 pKa = 4.49TNFKK94 pKa = 8.5FCKK97 pKa = 9.62WLTSPVAKK105 pKa = 10.29KK106 pKa = 10.45KK107 pKa = 10.21IAALQNADD115 pKa = 3.56RR116 pKa = 11.84LRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84GSDD123 pKa = 3.08VVTPEE128 pKa = 3.75EE129 pKa = 3.76AAMIALLEE137 pKa = 4.21SAQADD142 pKa = 3.97RR143 pKa = 11.84TTEE146 pKa = 3.86YY147 pKa = 10.44SKK149 pKa = 11.34ARR151 pKa = 11.84ARR153 pKa = 11.84YY154 pKa = 9.35DD155 pKa = 3.14IDD157 pKa = 3.37VAKK160 pKa = 10.4YY161 pKa = 9.17KK162 pKa = 10.79KK163 pKa = 10.3KK164 pKa = 8.77IAKK167 pKa = 7.74RR168 pKa = 11.84TRR170 pKa = 11.84QLEE173 pKa = 3.88EE174 pKa = 5.19DD175 pKa = 3.74LDD177 pKa = 4.27KK178 pKa = 11.69VQASYY183 pKa = 10.83PGLQLIEE190 pKa = 4.79RR191 pKa = 11.84PDD193 pKa = 3.62EE194 pKa = 4.36HH195 pKa = 9.05SVMASAWHH203 pKa = 6.72RR204 pKa = 11.84YY205 pKa = 7.58VDD207 pKa = 4.67FCTSNNFEE215 pKa = 4.29VPQKK219 pKa = 11.16NDD221 pKa = 2.85GNLARR226 pKa = 11.84AYY228 pKa = 9.69KK229 pKa = 9.92QFEE232 pKa = 4.26RR233 pKa = 11.84EE234 pKa = 3.8LTLEE238 pKa = 4.29IKK240 pKa = 9.21NTACQKK246 pKa = 10.61PEE248 pKa = 3.65VRR250 pKa = 11.84DD251 pKa = 3.66YY252 pKa = 11.57LLQYY256 pKa = 10.49CKK258 pKa = 10.72EE259 pKa = 3.98KK260 pKa = 11.17VKK262 pKa = 10.62GFRR265 pKa = 11.84AQFEE269 pKa = 4.55DD270 pKa = 4.96KK271 pKa = 10.56RR272 pKa = 11.84AQTYY276 pKa = 9.35RR277 pKa = 11.84GYY279 pKa = 9.81YY280 pKa = 9.83ACATGDD286 pKa = 3.98ILPEE290 pKa = 3.73DD291 pKa = 4.05TPRR294 pKa = 11.84KK295 pKa = 8.88KK296 pKa = 10.46RR297 pKa = 11.84KK298 pKa = 9.53ASRR301 pKa = 11.84PDD303 pKa = 3.05TGGEE307 pKa = 3.91ANQEE311 pKa = 4.06GEE313 pKa = 4.52DD314 pKa = 3.95DD315 pKa = 4.39SEE317 pKa = 4.94HH318 pKa = 7.56SVIPNLLDD326 pKa = 3.24TCVGDD331 pKa = 6.28DD332 pKa = 3.34RR333 pKa = 11.84RR334 pKa = 11.84AWDD337 pKa = 3.63KK338 pKa = 8.73THH340 pKa = 6.67PTNPEE345 pKa = 3.58KK346 pKa = 10.61PGNDD350 pKa = 3.23SSSDD354 pKa = 3.26EE355 pKa = 4.24SAEE358 pKa = 3.98NQPASTVADD367 pKa = 3.8RR368 pKa = 11.84TRR370 pKa = 11.84TRR372 pKa = 11.84RR373 pKa = 11.84RR374 pKa = 11.84KK375 pKa = 9.68ADD377 pKa = 3.23KK378 pKa = 10.29HH379 pKa = 5.83SKK381 pKa = 9.49KK382 pKa = 10.73
Molecular weight: 43.43 kDa
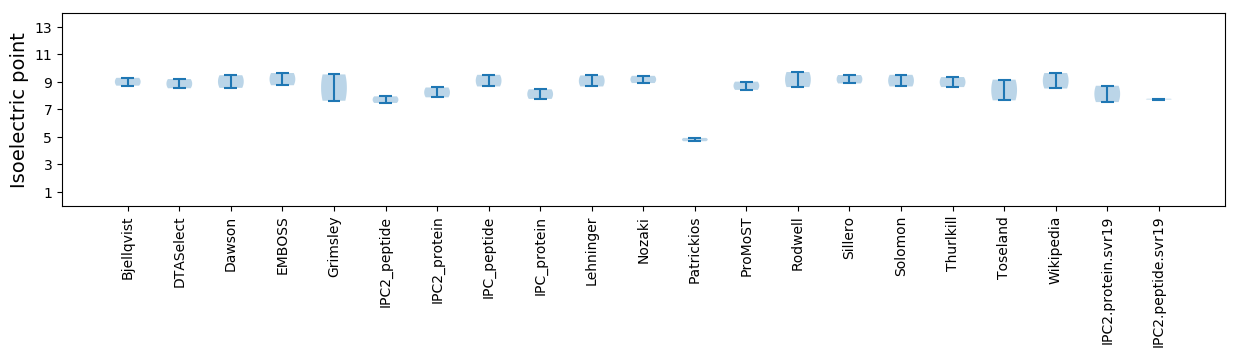
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6R5F0|A0A1W6R5F0_9VIRU Putative coat protein OS=Zostera marina amalgavirus 1 OX=1985202 PE=4 SV=1
MM1 pKa = 7.86AEE3 pKa = 4.71DD4 pKa = 4.62LRR6 pKa = 11.84SQQTLDD12 pKa = 4.41DD13 pKa = 4.27RR14 pKa = 11.84DD15 pKa = 3.9NLKK18 pKa = 10.9LLTDD22 pKa = 4.08AFKK25 pKa = 10.6AYY27 pKa = 9.65PEE29 pKa = 4.95GGLSVVDD36 pKa = 4.12VSLEE40 pKa = 4.15GIAACNYY47 pKa = 6.33TVSRR51 pKa = 11.84AVKK54 pKa = 9.85VMKK57 pKa = 10.04ILKK60 pKa = 9.25PLTKK64 pKa = 10.03NEE66 pKa = 3.86HH67 pKa = 6.22LIKK70 pKa = 10.47LFHH73 pKa = 6.38YY74 pKa = 10.79ANDD77 pKa = 3.25VSNISSILPMTLEE90 pKa = 4.49TNFKK94 pKa = 8.5FCKK97 pKa = 9.62WLTSPVAKK105 pKa = 10.29KK106 pKa = 10.45KK107 pKa = 10.21IAALQNADD115 pKa = 3.56RR116 pKa = 11.84LRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84GSDD123 pKa = 3.08VVTPEE128 pKa = 3.75EE129 pKa = 3.76AAMIALLEE137 pKa = 4.21SAQADD142 pKa = 3.97RR143 pKa = 11.84TTEE146 pKa = 3.86YY147 pKa = 10.44SKK149 pKa = 11.34ARR151 pKa = 11.84ARR153 pKa = 11.84YY154 pKa = 9.35DD155 pKa = 3.14IDD157 pKa = 3.37VAKK160 pKa = 10.4YY161 pKa = 9.17KK162 pKa = 10.79KK163 pKa = 10.3KK164 pKa = 8.77IAKK167 pKa = 7.74RR168 pKa = 11.84TRR170 pKa = 11.84QLEE173 pKa = 3.88EE174 pKa = 5.19DD175 pKa = 3.74LDD177 pKa = 4.27KK178 pKa = 11.69VQASYY183 pKa = 10.83PGLQLIEE190 pKa = 4.79RR191 pKa = 11.84PDD193 pKa = 3.62EE194 pKa = 4.36HH195 pKa = 9.05SVMASAWHH203 pKa = 6.72RR204 pKa = 11.84YY205 pKa = 7.58VDD207 pKa = 4.67FCTSNNFEE215 pKa = 4.29VPQKK219 pKa = 11.16NDD221 pKa = 2.85GNLARR226 pKa = 11.84AYY228 pKa = 9.69KK229 pKa = 9.92QFEE232 pKa = 4.26RR233 pKa = 11.84EE234 pKa = 3.8LTLEE238 pKa = 4.29IKK240 pKa = 9.21NTACQKK246 pKa = 10.61PEE248 pKa = 3.65VRR250 pKa = 11.84DD251 pKa = 3.66YY252 pKa = 11.57LLQYY256 pKa = 10.49CKK258 pKa = 10.6EE259 pKa = 4.05KK260 pKa = 11.29VKK262 pKa = 10.93GFVPNSKK269 pKa = 8.94TKK271 pKa = 8.86EE272 pKa = 3.87LKK274 pKa = 10.89LIGDD278 pKa = 3.96TMHH281 pKa = 7.02ARR283 pKa = 11.84LEE285 pKa = 4.54TYY287 pKa = 10.11FLRR290 pKa = 11.84IPLEE294 pKa = 3.79RR295 pKa = 11.84RR296 pKa = 11.84GRR298 pKa = 11.84LLDD301 pKa = 3.73QIPVGRR307 pKa = 11.84LTRR310 pKa = 11.84KK311 pKa = 10.23GKK313 pKa = 6.49MTPNIPLSQIFSTPALGMTAVHH335 pKa = 6.04GTKK338 pKa = 8.39PTPPIQRR345 pKa = 11.84NLEE348 pKa = 4.04MTLLPMNLLKK358 pKa = 10.37TNLRR362 pKa = 11.84VQLLTEE368 pKa = 4.2HH369 pKa = 5.92VRR371 pKa = 11.84GGVRR375 pKa = 11.84QISIARR381 pKa = 11.84SKK383 pKa = 10.1WEE385 pKa = 3.86AGVRR389 pKa = 11.84RR390 pKa = 11.84IIGGGEE396 pKa = 3.5MRR398 pKa = 11.84GWEE401 pKa = 4.42KK402 pKa = 11.24DD403 pKa = 3.2SALYY407 pKa = 10.43RR408 pKa = 11.84GGGNLHH414 pKa = 6.71DD415 pKa = 6.08AIRR418 pKa = 11.84LLATGRR424 pKa = 11.84QDD426 pKa = 3.31PPGSFLFEE434 pKa = 4.18HH435 pKa = 6.93FSLEE439 pKa = 4.04TARR442 pKa = 11.84EE443 pKa = 3.86ILLLPCNLEE452 pKa = 4.14VPDD455 pKa = 4.27GPKK458 pKa = 10.07AVRR461 pKa = 11.84MKK463 pKa = 10.91NFNEE467 pKa = 4.48DD468 pKa = 2.94ATAGPVLRR476 pKa = 11.84ALRR479 pKa = 11.84CKK481 pKa = 10.31SKK483 pKa = 11.09YY484 pKa = 9.35GLKK487 pKa = 10.48EE488 pKa = 3.64GLEE491 pKa = 4.23RR492 pKa = 11.84IAWDD496 pKa = 3.67LYY498 pKa = 11.48DD499 pKa = 3.92RR500 pKa = 11.84VGDD503 pKa = 3.65GKK505 pKa = 10.63LRR507 pKa = 11.84RR508 pKa = 11.84WQLPPLLARR517 pKa = 11.84IGFRR521 pKa = 11.84SKK523 pKa = 10.74LVEE526 pKa = 3.83QEE528 pKa = 4.1SAIKK532 pKa = 10.25KK533 pKa = 9.45IFSGQPIGRR542 pKa = 11.84AVMMLDD548 pKa = 4.01AMEE551 pKa = 4.57QPFSSPLYY559 pKa = 10.05NALCDD564 pKa = 3.81VVSKK568 pKa = 10.59LNRR571 pKa = 11.84VPEE574 pKa = 4.38SGWRR578 pKa = 11.84NMVVRR583 pKa = 11.84ASSDD587 pKa = 2.51WSRR590 pKa = 11.84FWGDD594 pKa = 2.93IRR596 pKa = 11.84NSCAIVEE603 pKa = 4.32LDD605 pKa = 2.84WSKK608 pKa = 11.02FDD610 pKa = 4.12RR611 pKa = 11.84EE612 pKa = 4.14RR613 pKa = 11.84PKK615 pKa = 11.03EE616 pKa = 4.32DD617 pKa = 2.98IEE619 pKa = 4.16FVINIFVSCFLPKK632 pKa = 10.04NRR634 pKa = 11.84RR635 pKa = 11.84EE636 pKa = 3.88RR637 pKa = 11.84RR638 pKa = 11.84LLSAYY643 pKa = 9.69KK644 pKa = 10.68YY645 pKa = 9.71MMRR648 pKa = 11.84KK649 pKa = 9.51ALIDD653 pKa = 3.74KK654 pKa = 10.73VIMLDD659 pKa = 3.53DD660 pKa = 4.38GCAFTYY666 pKa = 10.8DD667 pKa = 3.34GMIPSGSLWTGLLGTALNILYY688 pKa = 10.19INAAVRR694 pKa = 11.84RR695 pKa = 11.84LGIPSDD701 pKa = 3.7EE702 pKa = 4.15FVPYY706 pKa = 10.42CAGDD710 pKa = 3.79DD711 pKa = 3.57NLTVFKK717 pKa = 10.63YY718 pKa = 9.06PQRR721 pKa = 11.84ATRR724 pKa = 11.84LGKK727 pKa = 9.78IRR729 pKa = 11.84GMLNEE734 pKa = 4.34MFRR737 pKa = 11.84AGIDD741 pKa = 3.39PEE743 pKa = 4.75DD744 pKa = 4.96FIIHH748 pKa = 5.99YY749 pKa = 9.89PPFHH753 pKa = 5.4VTKK756 pKa = 10.5VQAKK760 pKa = 8.74FPAGFDD766 pKa = 3.73FTHH769 pKa = 6.37GTSKK773 pKa = 10.72FLDD776 pKa = 3.37QCQWIPLKK784 pKa = 11.25GNIHH788 pKa = 5.99VDD790 pKa = 3.03HH791 pKa = 6.99SEE793 pKa = 4.12GLSHH797 pKa = 7.11RR798 pKa = 11.84WNYY801 pKa = 8.62VFKK804 pKa = 10.94GKK806 pKa = 9.64PKK808 pKa = 10.08FLANYY813 pKa = 9.2FMEE816 pKa = 5.62DD817 pKa = 3.09GRR819 pKa = 11.84SIRR822 pKa = 11.84PAHH825 pKa = 6.99DD826 pKa = 3.57NLEE829 pKa = 4.12KK830 pKa = 10.79LLYY833 pKa = 9.45PEE835 pKa = 5.23NVQQKK840 pKa = 9.77IEE842 pKa = 4.26DD843 pKa = 3.69YY844 pKa = 9.31EE845 pKa = 4.23AAVLSMAVDD854 pKa = 4.03NPWNSHH860 pKa = 5.81NINHH864 pKa = 6.62LMQRR868 pKa = 11.84FCIIQQIKK876 pKa = 8.86RR877 pKa = 11.84QAVHH881 pKa = 7.18PLTAGDD887 pKa = 4.06VLFCAKK893 pKa = 9.85FQSISEE899 pKa = 4.23NMWMFPTVGAWRR911 pKa = 11.84RR912 pKa = 11.84QLLNVRR918 pKa = 11.84MEE920 pKa = 5.35DD921 pKa = 3.49IEE923 pKa = 4.21EE924 pKa = 4.22LKK926 pKa = 9.61PTVEE930 pKa = 4.11AFSEE934 pKa = 4.7FVRR937 pKa = 11.84GVTSLYY943 pKa = 10.86SRR945 pKa = 11.84ASTGGLDD952 pKa = 2.73SWQFMEE958 pKa = 4.18ILRR961 pKa = 11.84GEE963 pKa = 4.07RR964 pKa = 11.84DD965 pKa = 3.48AGSGQYY971 pKa = 10.92GSDD974 pKa = 3.09IEE976 pKa = 4.27AWCAFLNRR984 pKa = 11.84NGLTRR989 pKa = 11.84SLRR992 pKa = 11.84PAKK995 pKa = 10.15RR996 pKa = 11.84YY997 pKa = 9.64RR998 pKa = 11.84KK999 pKa = 9.29EE1000 pKa = 3.62KK1001 pKa = 9.1QTYY1004 pKa = 8.42QSAEE1008 pKa = 3.99AGDD1011 pKa = 4.46KK1012 pKa = 10.82ILVSLAAIYY1021 pKa = 7.63ATSDD1025 pKa = 2.92YY1026 pKa = 11.23DD1027 pKa = 3.96LRR1029 pKa = 11.84EE1030 pKa = 4.24DD1031 pKa = 4.02RR1032 pKa = 11.84DD1033 pKa = 3.54LGFIIRR1039 pKa = 11.84ISNEE1043 pKa = 3.24LRR1045 pKa = 11.84KK1046 pKa = 9.61QLLL1049 pKa = 3.44
MM1 pKa = 7.86AEE3 pKa = 4.71DD4 pKa = 4.62LRR6 pKa = 11.84SQQTLDD12 pKa = 4.41DD13 pKa = 4.27RR14 pKa = 11.84DD15 pKa = 3.9NLKK18 pKa = 10.9LLTDD22 pKa = 4.08AFKK25 pKa = 10.6AYY27 pKa = 9.65PEE29 pKa = 4.95GGLSVVDD36 pKa = 4.12VSLEE40 pKa = 4.15GIAACNYY47 pKa = 6.33TVSRR51 pKa = 11.84AVKK54 pKa = 9.85VMKK57 pKa = 10.04ILKK60 pKa = 9.25PLTKK64 pKa = 10.03NEE66 pKa = 3.86HH67 pKa = 6.22LIKK70 pKa = 10.47LFHH73 pKa = 6.38YY74 pKa = 10.79ANDD77 pKa = 3.25VSNISSILPMTLEE90 pKa = 4.49TNFKK94 pKa = 8.5FCKK97 pKa = 9.62WLTSPVAKK105 pKa = 10.29KK106 pKa = 10.45KK107 pKa = 10.21IAALQNADD115 pKa = 3.56RR116 pKa = 11.84LRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84GSDD123 pKa = 3.08VVTPEE128 pKa = 3.75EE129 pKa = 3.76AAMIALLEE137 pKa = 4.21SAQADD142 pKa = 3.97RR143 pKa = 11.84TTEE146 pKa = 3.86YY147 pKa = 10.44SKK149 pKa = 11.34ARR151 pKa = 11.84ARR153 pKa = 11.84YY154 pKa = 9.35DD155 pKa = 3.14IDD157 pKa = 3.37VAKK160 pKa = 10.4YY161 pKa = 9.17KK162 pKa = 10.79KK163 pKa = 10.3KK164 pKa = 8.77IAKK167 pKa = 7.74RR168 pKa = 11.84TRR170 pKa = 11.84QLEE173 pKa = 3.88EE174 pKa = 5.19DD175 pKa = 3.74LDD177 pKa = 4.27KK178 pKa = 11.69VQASYY183 pKa = 10.83PGLQLIEE190 pKa = 4.79RR191 pKa = 11.84PDD193 pKa = 3.62EE194 pKa = 4.36HH195 pKa = 9.05SVMASAWHH203 pKa = 6.72RR204 pKa = 11.84YY205 pKa = 7.58VDD207 pKa = 4.67FCTSNNFEE215 pKa = 4.29VPQKK219 pKa = 11.16NDD221 pKa = 2.85GNLARR226 pKa = 11.84AYY228 pKa = 9.69KK229 pKa = 9.92QFEE232 pKa = 4.26RR233 pKa = 11.84EE234 pKa = 3.8LTLEE238 pKa = 4.29IKK240 pKa = 9.21NTACQKK246 pKa = 10.61PEE248 pKa = 3.65VRR250 pKa = 11.84DD251 pKa = 3.66YY252 pKa = 11.57LLQYY256 pKa = 10.49CKK258 pKa = 10.6EE259 pKa = 4.05KK260 pKa = 11.29VKK262 pKa = 10.93GFVPNSKK269 pKa = 8.94TKK271 pKa = 8.86EE272 pKa = 3.87LKK274 pKa = 10.89LIGDD278 pKa = 3.96TMHH281 pKa = 7.02ARR283 pKa = 11.84LEE285 pKa = 4.54TYY287 pKa = 10.11FLRR290 pKa = 11.84IPLEE294 pKa = 3.79RR295 pKa = 11.84RR296 pKa = 11.84GRR298 pKa = 11.84LLDD301 pKa = 3.73QIPVGRR307 pKa = 11.84LTRR310 pKa = 11.84KK311 pKa = 10.23GKK313 pKa = 6.49MTPNIPLSQIFSTPALGMTAVHH335 pKa = 6.04GTKK338 pKa = 8.39PTPPIQRR345 pKa = 11.84NLEE348 pKa = 4.04MTLLPMNLLKK358 pKa = 10.37TNLRR362 pKa = 11.84VQLLTEE368 pKa = 4.2HH369 pKa = 5.92VRR371 pKa = 11.84GGVRR375 pKa = 11.84QISIARR381 pKa = 11.84SKK383 pKa = 10.1WEE385 pKa = 3.86AGVRR389 pKa = 11.84RR390 pKa = 11.84IIGGGEE396 pKa = 3.5MRR398 pKa = 11.84GWEE401 pKa = 4.42KK402 pKa = 11.24DD403 pKa = 3.2SALYY407 pKa = 10.43RR408 pKa = 11.84GGGNLHH414 pKa = 6.71DD415 pKa = 6.08AIRR418 pKa = 11.84LLATGRR424 pKa = 11.84QDD426 pKa = 3.31PPGSFLFEE434 pKa = 4.18HH435 pKa = 6.93FSLEE439 pKa = 4.04TARR442 pKa = 11.84EE443 pKa = 3.86ILLLPCNLEE452 pKa = 4.14VPDD455 pKa = 4.27GPKK458 pKa = 10.07AVRR461 pKa = 11.84MKK463 pKa = 10.91NFNEE467 pKa = 4.48DD468 pKa = 2.94ATAGPVLRR476 pKa = 11.84ALRR479 pKa = 11.84CKK481 pKa = 10.31SKK483 pKa = 11.09YY484 pKa = 9.35GLKK487 pKa = 10.48EE488 pKa = 3.64GLEE491 pKa = 4.23RR492 pKa = 11.84IAWDD496 pKa = 3.67LYY498 pKa = 11.48DD499 pKa = 3.92RR500 pKa = 11.84VGDD503 pKa = 3.65GKK505 pKa = 10.63LRR507 pKa = 11.84RR508 pKa = 11.84WQLPPLLARR517 pKa = 11.84IGFRR521 pKa = 11.84SKK523 pKa = 10.74LVEE526 pKa = 3.83QEE528 pKa = 4.1SAIKK532 pKa = 10.25KK533 pKa = 9.45IFSGQPIGRR542 pKa = 11.84AVMMLDD548 pKa = 4.01AMEE551 pKa = 4.57QPFSSPLYY559 pKa = 10.05NALCDD564 pKa = 3.81VVSKK568 pKa = 10.59LNRR571 pKa = 11.84VPEE574 pKa = 4.38SGWRR578 pKa = 11.84NMVVRR583 pKa = 11.84ASSDD587 pKa = 2.51WSRR590 pKa = 11.84FWGDD594 pKa = 2.93IRR596 pKa = 11.84NSCAIVEE603 pKa = 4.32LDD605 pKa = 2.84WSKK608 pKa = 11.02FDD610 pKa = 4.12RR611 pKa = 11.84EE612 pKa = 4.14RR613 pKa = 11.84PKK615 pKa = 11.03EE616 pKa = 4.32DD617 pKa = 2.98IEE619 pKa = 4.16FVINIFVSCFLPKK632 pKa = 10.04NRR634 pKa = 11.84RR635 pKa = 11.84EE636 pKa = 3.88RR637 pKa = 11.84RR638 pKa = 11.84LLSAYY643 pKa = 9.69KK644 pKa = 10.68YY645 pKa = 9.71MMRR648 pKa = 11.84KK649 pKa = 9.51ALIDD653 pKa = 3.74KK654 pKa = 10.73VIMLDD659 pKa = 3.53DD660 pKa = 4.38GCAFTYY666 pKa = 10.8DD667 pKa = 3.34GMIPSGSLWTGLLGTALNILYY688 pKa = 10.19INAAVRR694 pKa = 11.84RR695 pKa = 11.84LGIPSDD701 pKa = 3.7EE702 pKa = 4.15FVPYY706 pKa = 10.42CAGDD710 pKa = 3.79DD711 pKa = 3.57NLTVFKK717 pKa = 10.63YY718 pKa = 9.06PQRR721 pKa = 11.84ATRR724 pKa = 11.84LGKK727 pKa = 9.78IRR729 pKa = 11.84GMLNEE734 pKa = 4.34MFRR737 pKa = 11.84AGIDD741 pKa = 3.39PEE743 pKa = 4.75DD744 pKa = 4.96FIIHH748 pKa = 5.99YY749 pKa = 9.89PPFHH753 pKa = 5.4VTKK756 pKa = 10.5VQAKK760 pKa = 8.74FPAGFDD766 pKa = 3.73FTHH769 pKa = 6.37GTSKK773 pKa = 10.72FLDD776 pKa = 3.37QCQWIPLKK784 pKa = 11.25GNIHH788 pKa = 5.99VDD790 pKa = 3.03HH791 pKa = 6.99SEE793 pKa = 4.12GLSHH797 pKa = 7.11RR798 pKa = 11.84WNYY801 pKa = 8.62VFKK804 pKa = 10.94GKK806 pKa = 9.64PKK808 pKa = 10.08FLANYY813 pKa = 9.2FMEE816 pKa = 5.62DD817 pKa = 3.09GRR819 pKa = 11.84SIRR822 pKa = 11.84PAHH825 pKa = 6.99DD826 pKa = 3.57NLEE829 pKa = 4.12KK830 pKa = 10.79LLYY833 pKa = 9.45PEE835 pKa = 5.23NVQQKK840 pKa = 9.77IEE842 pKa = 4.26DD843 pKa = 3.69YY844 pKa = 9.31EE845 pKa = 4.23AAVLSMAVDD854 pKa = 4.03NPWNSHH860 pKa = 5.81NINHH864 pKa = 6.62LMQRR868 pKa = 11.84FCIIQQIKK876 pKa = 8.86RR877 pKa = 11.84QAVHH881 pKa = 7.18PLTAGDD887 pKa = 4.06VLFCAKK893 pKa = 9.85FQSISEE899 pKa = 4.23NMWMFPTVGAWRR911 pKa = 11.84RR912 pKa = 11.84QLLNVRR918 pKa = 11.84MEE920 pKa = 5.35DD921 pKa = 3.49IEE923 pKa = 4.21EE924 pKa = 4.22LKK926 pKa = 9.61PTVEE930 pKa = 4.11AFSEE934 pKa = 4.7FVRR937 pKa = 11.84GVTSLYY943 pKa = 10.86SRR945 pKa = 11.84ASTGGLDD952 pKa = 2.73SWQFMEE958 pKa = 4.18ILRR961 pKa = 11.84GEE963 pKa = 4.07RR964 pKa = 11.84DD965 pKa = 3.48AGSGQYY971 pKa = 10.92GSDD974 pKa = 3.09IEE976 pKa = 4.27AWCAFLNRR984 pKa = 11.84NGLTRR989 pKa = 11.84SLRR992 pKa = 11.84PAKK995 pKa = 10.15RR996 pKa = 11.84YY997 pKa = 9.64RR998 pKa = 11.84KK999 pKa = 9.29EE1000 pKa = 3.62KK1001 pKa = 9.1QTYY1004 pKa = 8.42QSAEE1008 pKa = 3.99AGDD1011 pKa = 4.46KK1012 pKa = 10.82ILVSLAAIYY1021 pKa = 7.63ATSDD1025 pKa = 2.92YY1026 pKa = 11.23DD1027 pKa = 3.96LRR1029 pKa = 11.84EE1030 pKa = 4.24DD1031 pKa = 4.02RR1032 pKa = 11.84DD1033 pKa = 3.54LGFIIRR1039 pKa = 11.84ISNEE1043 pKa = 3.24LRR1045 pKa = 11.84KK1046 pKa = 9.61QLLL1049 pKa = 3.44
Molecular weight: 119.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1431 |
382 |
1049 |
715.5 |
81.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.036 ± 0.904 | 1.607 ± 0.123 |
6.499 ± 1.03 | 6.499 ± 0.456 |
3.704 ± 0.739 | 5.451 ± 0.979 |
1.817 ± 0.009 | 5.101 ± 0.931 |
7.407 ± 1.106 | 9.853 ± 1.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.376 ± 0.585 | 4.333 ± 0.064 |
4.682 ± 0.127 | 3.704 ± 0.122 |
8.036 ± 0.1 | 5.94 ± 0.188 |
4.822 ± 0.801 | 5.381 ± 0.08 |
1.468 ± 0.374 | 3.284 ± 0.209 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
