
Ixeridium yellow mottle virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Calvusvirinae; Umbravirus
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
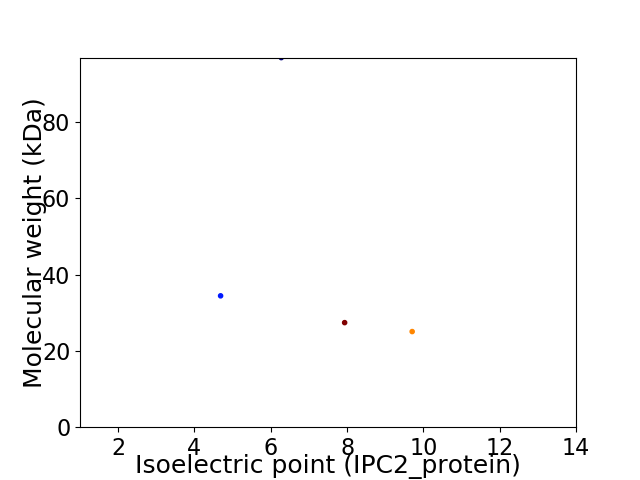
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U8YLV2|A0A1U8YLV2_9TOMB Putative movement protein OS=Ixeridium yellow mottle virus 2 OX=1817526 GN=orf4 PE=4 SV=1
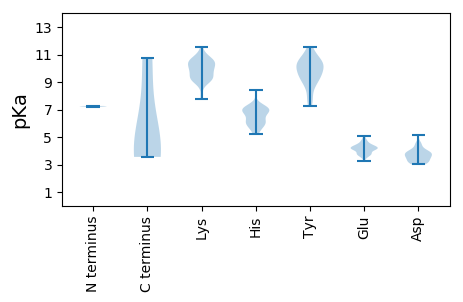
MM1 pKa = 7.23VRR3 pKa = 11.84FADD6 pKa = 4.21EE7 pKa = 3.71EE8 pKa = 4.25WALLVCQQRR17 pKa = 11.84MTDD20 pKa = 3.1AHH22 pKa = 5.92QATLNEE28 pKa = 4.42WYY30 pKa = 9.59EE31 pKa = 4.14GTIEE35 pKa = 4.13TDD37 pKa = 3.22VYY39 pKa = 10.83IPLSGEE45 pKa = 3.72PTNACEE51 pKa = 4.33ALPVAEE57 pKa = 4.34PQVGPVAIPLDD68 pKa = 4.02GSHH71 pKa = 7.13RR72 pKa = 11.84STVDD76 pKa = 3.01ALAGSEE82 pKa = 4.13DD83 pKa = 3.84APGVSGDD90 pKa = 3.73ASSTTSLEE98 pKa = 4.37TYY100 pKa = 10.32LGAAHH105 pKa = 7.06EE106 pKa = 4.14LLKK109 pKa = 10.57IEE111 pKa = 4.66AEE113 pKa = 4.1IAGLQLPQTGEE124 pKa = 3.85EE125 pKa = 4.11EE126 pKa = 4.77AYY128 pKa = 10.38GPHH131 pKa = 6.28YY132 pKa = 10.24PSGYY136 pKa = 9.91CNNPCRR142 pKa = 11.84ALVPWQPLVIKK153 pKa = 9.95VPRR156 pKa = 11.84APAPVIYY163 pKa = 10.27CGYY166 pKa = 7.6TPGWEE171 pKa = 4.19EE172 pKa = 3.54SRR174 pKa = 11.84SPVEE178 pKa = 3.73RR179 pKa = 11.84LGAAVVEE186 pKa = 4.58TGNLLAGYY194 pKa = 9.44LFTKK198 pKa = 9.31VIPFAAGTVFHH209 pKa = 7.14SMAQLATMVAEE220 pKa = 5.06AEE222 pKa = 4.48QPCDD226 pKa = 3.5PTIEE230 pKa = 4.73GPTLPLEE237 pKa = 4.34EE238 pKa = 4.87RR239 pKa = 11.84GAAMGYY245 pKa = 8.72MNSAAIAMEE254 pKa = 3.86LRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84FGRR261 pKa = 11.84PQATPANVEE270 pKa = 3.69LGGRR274 pKa = 11.84VARR277 pKa = 11.84EE278 pKa = 3.73ILSDD282 pKa = 3.55RR283 pKa = 11.84CRR285 pKa = 11.84ATRR288 pKa = 11.84SEE290 pKa = 3.58AWYY293 pKa = 8.87TSVEE297 pKa = 4.04ATKK300 pKa = 9.36MWLTPTLVDD309 pKa = 3.58MVHH312 pKa = 5.89ATGNVGFCC320 pKa = 4.19
MM1 pKa = 7.23VRR3 pKa = 11.84FADD6 pKa = 4.21EE7 pKa = 3.71EE8 pKa = 4.25WALLVCQQRR17 pKa = 11.84MTDD20 pKa = 3.1AHH22 pKa = 5.92QATLNEE28 pKa = 4.42WYY30 pKa = 9.59EE31 pKa = 4.14GTIEE35 pKa = 4.13TDD37 pKa = 3.22VYY39 pKa = 10.83IPLSGEE45 pKa = 3.72PTNACEE51 pKa = 4.33ALPVAEE57 pKa = 4.34PQVGPVAIPLDD68 pKa = 4.02GSHH71 pKa = 7.13RR72 pKa = 11.84STVDD76 pKa = 3.01ALAGSEE82 pKa = 4.13DD83 pKa = 3.84APGVSGDD90 pKa = 3.73ASSTTSLEE98 pKa = 4.37TYY100 pKa = 10.32LGAAHH105 pKa = 7.06EE106 pKa = 4.14LLKK109 pKa = 10.57IEE111 pKa = 4.66AEE113 pKa = 4.1IAGLQLPQTGEE124 pKa = 3.85EE125 pKa = 4.11EE126 pKa = 4.77AYY128 pKa = 10.38GPHH131 pKa = 6.28YY132 pKa = 10.24PSGYY136 pKa = 9.91CNNPCRR142 pKa = 11.84ALVPWQPLVIKK153 pKa = 9.95VPRR156 pKa = 11.84APAPVIYY163 pKa = 10.27CGYY166 pKa = 7.6TPGWEE171 pKa = 4.19EE172 pKa = 3.54SRR174 pKa = 11.84SPVEE178 pKa = 3.73RR179 pKa = 11.84LGAAVVEE186 pKa = 4.58TGNLLAGYY194 pKa = 9.44LFTKK198 pKa = 9.31VIPFAAGTVFHH209 pKa = 7.14SMAQLATMVAEE220 pKa = 5.06AEE222 pKa = 4.48QPCDD226 pKa = 3.5PTIEE230 pKa = 4.73GPTLPLEE237 pKa = 4.34EE238 pKa = 4.87RR239 pKa = 11.84GAAMGYY245 pKa = 8.72MNSAAIAMEE254 pKa = 3.86LRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84FGRR261 pKa = 11.84PQATPANVEE270 pKa = 3.69LGGRR274 pKa = 11.84VARR277 pKa = 11.84EE278 pKa = 3.73ILSDD282 pKa = 3.55RR283 pKa = 11.84CRR285 pKa = 11.84ATRR288 pKa = 11.84SEE290 pKa = 3.58AWYY293 pKa = 8.87TSVEE297 pKa = 4.04ATKK300 pKa = 9.36MWLTPTLVDD309 pKa = 3.58MVHH312 pKa = 5.89ATGNVGFCC320 pKa = 4.19
Molecular weight: 34.45 kDa
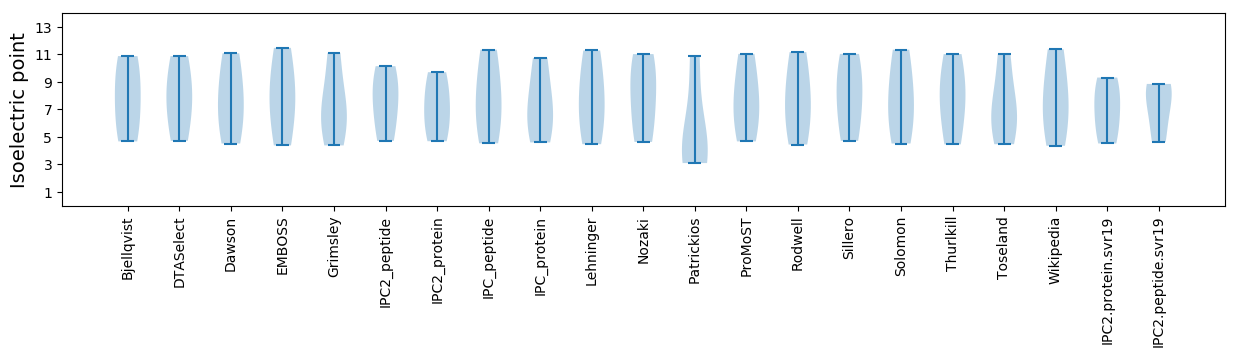
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U8YMV9|A0A1U8YMV9_9TOMB RNA-directed RNA polymerase OS=Ixeridium yellow mottle virus 2 OX=1817526 GN=IYMV_gp1 PE=4 SV=1
MM1 pKa = 7.26TNVFNVYY8 pKa = 9.45TPDD11 pKa = 3.17TSKK14 pKa = 10.7QPRR17 pKa = 11.84GASANAGRR25 pKa = 11.84RR26 pKa = 11.84PPTGVGRR33 pKa = 11.84LWDD36 pKa = 3.8GHH38 pKa = 5.19NHH40 pKa = 5.63PGARR44 pKa = 11.84KK45 pKa = 9.19RR46 pKa = 11.84EE47 pKa = 4.5SNVRR51 pKa = 11.84TPPAPKK57 pKa = 10.23GPQDD61 pKa = 3.44HH62 pKa = 7.14AGQATHH68 pKa = 7.07AKK70 pKa = 9.79GNPKK74 pKa = 9.67HH75 pKa = 6.66RR76 pKa = 11.84GPVVHH81 pKa = 6.88PAAGGGVRR89 pKa = 11.84PTRR92 pKa = 11.84PRR94 pKa = 11.84RR95 pKa = 11.84LPRR98 pKa = 11.84DD99 pKa = 3.75GEE101 pKa = 4.28PLDD104 pKa = 4.51HH105 pKa = 7.34RR106 pKa = 11.84SNPAQRR112 pKa = 11.84KK113 pKa = 8.0RR114 pKa = 11.84RR115 pKa = 11.84VRR117 pKa = 11.84KK118 pKa = 9.85SKK120 pKa = 9.13LTPEE124 pKa = 3.47QGAAIRR130 pKa = 11.84ALSATLFDD138 pKa = 3.82TLIGYY143 pKa = 7.26GVRR146 pKa = 11.84KK147 pKa = 9.38GALLQHH153 pKa = 6.73CLRR156 pKa = 11.84AVRR159 pKa = 11.84DD160 pKa = 3.71SHH162 pKa = 6.85SEE164 pKa = 3.25WRR166 pKa = 11.84KK167 pKa = 9.13RR168 pKa = 11.84LQSVSPVGARR178 pKa = 11.84GEE180 pKa = 4.14PSGSQLPSSTADD192 pKa = 3.11CKK194 pKa = 10.43TSPEE198 pKa = 4.29AGTIGPIEE206 pKa = 4.35KK207 pKa = 9.45PAAVEE212 pKa = 4.24GTSAAGDD219 pKa = 3.94DD220 pKa = 3.9QPTLCRR226 pKa = 11.84SCTGHH231 pKa = 6.15NNVYY235 pKa = 10.78
MM1 pKa = 7.26TNVFNVYY8 pKa = 9.45TPDD11 pKa = 3.17TSKK14 pKa = 10.7QPRR17 pKa = 11.84GASANAGRR25 pKa = 11.84RR26 pKa = 11.84PPTGVGRR33 pKa = 11.84LWDD36 pKa = 3.8GHH38 pKa = 5.19NHH40 pKa = 5.63PGARR44 pKa = 11.84KK45 pKa = 9.19RR46 pKa = 11.84EE47 pKa = 4.5SNVRR51 pKa = 11.84TPPAPKK57 pKa = 10.23GPQDD61 pKa = 3.44HH62 pKa = 7.14AGQATHH68 pKa = 7.07AKK70 pKa = 9.79GNPKK74 pKa = 9.67HH75 pKa = 6.66RR76 pKa = 11.84GPVVHH81 pKa = 6.88PAAGGGVRR89 pKa = 11.84PTRR92 pKa = 11.84PRR94 pKa = 11.84RR95 pKa = 11.84LPRR98 pKa = 11.84DD99 pKa = 3.75GEE101 pKa = 4.28PLDD104 pKa = 4.51HH105 pKa = 7.34RR106 pKa = 11.84SNPAQRR112 pKa = 11.84KK113 pKa = 8.0RR114 pKa = 11.84RR115 pKa = 11.84VRR117 pKa = 11.84KK118 pKa = 9.85SKK120 pKa = 9.13LTPEE124 pKa = 3.47QGAAIRR130 pKa = 11.84ALSATLFDD138 pKa = 3.82TLIGYY143 pKa = 7.26GVRR146 pKa = 11.84KK147 pKa = 9.38GALLQHH153 pKa = 6.73CLRR156 pKa = 11.84AVRR159 pKa = 11.84DD160 pKa = 3.71SHH162 pKa = 6.85SEE164 pKa = 3.25WRR166 pKa = 11.84KK167 pKa = 9.13RR168 pKa = 11.84LQSVSPVGARR178 pKa = 11.84GEE180 pKa = 4.14PSGSQLPSSTADD192 pKa = 3.11CKK194 pKa = 10.43TSPEE198 pKa = 4.29AGTIGPIEE206 pKa = 4.35KK207 pKa = 9.45PAAVEE212 pKa = 4.24GTSAAGDD219 pKa = 3.94DD220 pKa = 3.9QPTLCRR226 pKa = 11.84SCTGHH231 pKa = 6.15NNVYY235 pKa = 10.78
Molecular weight: 25.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1677 |
235 |
872 |
419.3 |
45.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.72 ± 0.925 | 2.326 ± 0.262 |
3.936 ± 0.22 | 6.261 ± 1.066 |
2.922 ± 0.698 | 7.99 ± 0.663 |
2.504 ± 0.355 | 4.055 ± 0.584 |
3.578 ± 0.775 | 8.766 ± 0.962 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.624 ± 0.509 | 3.518 ± 0.29 |
7.692 ± 0.785 | 3.637 ± 0.496 |
6.857 ± 0.933 | 5.307 ± 0.443 |
6.738 ± 0.219 | 7.275 ± 0.288 |
1.491 ± 0.224 | 2.803 ± 0.536 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
