
Anoxybacter fermentans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Halanaerobiales; Halanaerobiales incertae sedis; Anoxybacter
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
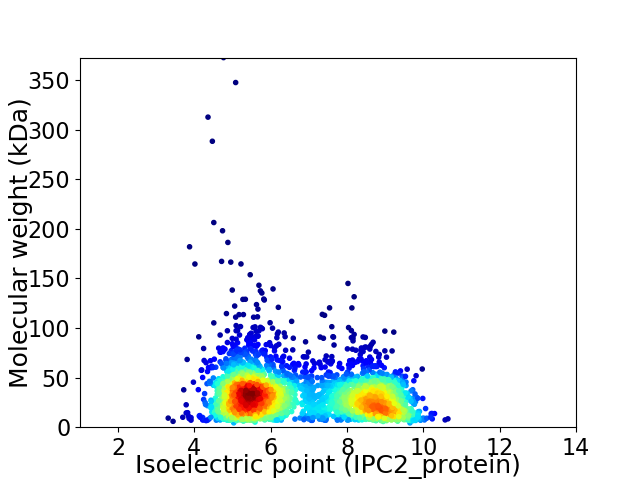
Virtual 2D-PAGE plot for 3011 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9HRZ4|A0A3Q9HRZ4_9FIRM Probable 2-phosphosulfolactate phosphatase OS=Anoxybacter fermentans OX=1323375 GN=comB PE=3 SV=1
MM1 pKa = 7.55KK2 pKa = 10.51KK3 pKa = 10.43LLTLLTILILTVGLVGCDD21 pKa = 3.68FLPFMNKK28 pKa = 9.9APVIYY33 pKa = 10.44SLISEE38 pKa = 4.67KK39 pKa = 8.31TTIHH43 pKa = 6.54PGEE46 pKa = 4.04EE47 pKa = 3.63VKK49 pKa = 10.72IQVKK53 pKa = 10.5ASDD56 pKa = 4.11PNGDD60 pKa = 3.7SLVYY64 pKa = 9.04TWNATGGKK72 pKa = 9.57INGTGSEE79 pKa = 4.7VIWVAPQEE87 pKa = 4.05IGEE90 pKa = 4.06YY91 pKa = 9.22TITVKK96 pKa = 10.73VSDD99 pKa = 4.13SKK101 pKa = 11.77VDD103 pKa = 3.43VSKK106 pKa = 10.98SIQISVQPLPNEE118 pKa = 4.02APVISSLTVEE128 pKa = 4.35KK129 pKa = 10.86SVVQPEE135 pKa = 4.42EE136 pKa = 3.89KK137 pKa = 10.75VKK139 pKa = 11.03VQVQASDD146 pKa = 3.61PDD148 pKa = 4.39GDD150 pKa = 3.81VLTYY154 pKa = 9.15TWTVTGGSIEE164 pKa = 4.31GTGSEE169 pKa = 4.52VFWIASQTDD178 pKa = 3.44GNYY181 pKa = 9.57TITVEE186 pKa = 4.16VSDD189 pKa = 4.86GRR191 pKa = 11.84DD192 pKa = 3.09TVTGSINIQVNTAPIIEE209 pKa = 5.01EE210 pKa = 3.85ISASSSSVLVNGTLTLTAIVNDD232 pKa = 3.8PTGDD236 pKa = 3.31NLTYY240 pKa = 10.15RR241 pKa = 11.84WEE243 pKa = 4.13DD244 pKa = 3.41AIGEE248 pKa = 4.31VLGTEE253 pKa = 5.07PSLEE257 pKa = 3.6WHH259 pKa = 6.92APDD262 pKa = 3.5TYY264 pKa = 11.02GVYY267 pKa = 10.27PVTLYY272 pKa = 11.12VSDD275 pKa = 5.4GIFEE279 pKa = 4.14SSKK282 pKa = 9.61TININVEE289 pKa = 4.3DD290 pKa = 4.84DD291 pKa = 3.46SHH293 pKa = 6.5HH294 pKa = 6.41APEE297 pKa = 4.21IQEE300 pKa = 4.18VAITTPYY307 pKa = 10.45FEE309 pKa = 4.4SQVVVDD315 pKa = 4.53SVLAAGRR322 pKa = 11.84LYY324 pKa = 10.68AVWPIFVDD332 pKa = 3.44VDD334 pKa = 3.59EE335 pKa = 5.47SDD337 pKa = 3.53QLDD340 pKa = 3.63IQATADD346 pKa = 3.4VGEE349 pKa = 4.32VEE351 pKa = 4.66IFASEE356 pKa = 4.51YY357 pKa = 9.27EE358 pKa = 4.63TICFWVAPDD367 pKa = 3.4VAGEE371 pKa = 4.02YY372 pKa = 10.07TLTFTVTDD380 pKa = 4.77GIYY383 pKa = 9.78TVVKK387 pKa = 8.94EE388 pKa = 4.19YY389 pKa = 10.31PVQVVEE395 pKa = 4.32NSCPEE400 pKa = 3.31IDD402 pKa = 4.91AIYY405 pKa = 10.34INNEE409 pKa = 4.35LITDD413 pKa = 4.34NYY415 pKa = 8.63YY416 pKa = 10.15QAAPGEE422 pKa = 4.52EE423 pKa = 3.76ITISVLASDD432 pKa = 4.96SDD434 pKa = 3.6NDD436 pKa = 3.57APLEE440 pKa = 3.94YY441 pKa = 10.39KK442 pKa = 10.35IIDD445 pKa = 3.93PYY447 pKa = 10.62YY448 pKa = 10.04EE449 pKa = 3.42IWNTNEE455 pKa = 3.91LTFTAPMEE463 pKa = 4.16EE464 pKa = 4.34EE465 pKa = 3.86MDD467 pKa = 4.37EE468 pKa = 3.53IWIYY472 pKa = 11.28VFDD475 pKa = 4.21GLQVRR480 pKa = 11.84VASVIIDD487 pKa = 3.57VDD489 pKa = 3.48NGADD493 pKa = 4.44NNTNDD498 pKa = 5.13QNAIDD503 pKa = 4.35QNAIDD508 pKa = 4.57QINNLISRR516 pKa = 11.84FVNDD520 pKa = 5.01YY521 pKa = 10.62EE522 pKa = 4.27NQDD525 pKa = 3.25INSILSYY532 pKa = 10.6TDD534 pKa = 3.27SSYY537 pKa = 10.98QAEE540 pKa = 4.47LKK542 pKa = 10.68SRR544 pKa = 11.84LNRR547 pKa = 11.84YY548 pKa = 9.18FEE550 pKa = 4.51LEE552 pKa = 3.98RR553 pKa = 11.84VSNLTIRR560 pKa = 11.84KK561 pKa = 8.43LSEE564 pKa = 3.96IIEE567 pKa = 4.09VDD569 pKa = 3.47GEE571 pKa = 4.45FEE573 pKa = 4.19VQVAVQGDD581 pKa = 4.4FYY583 pKa = 11.72VLFEE587 pKa = 4.35TFPNSLIRR595 pKa = 11.84VFTFWFTYY603 pKa = 10.5DD604 pKa = 2.93EE605 pKa = 4.48YY606 pKa = 11.79GNILISSVWLL616 pKa = 3.62
MM1 pKa = 7.55KK2 pKa = 10.51KK3 pKa = 10.43LLTLLTILILTVGLVGCDD21 pKa = 3.68FLPFMNKK28 pKa = 9.9APVIYY33 pKa = 10.44SLISEE38 pKa = 4.67KK39 pKa = 8.31TTIHH43 pKa = 6.54PGEE46 pKa = 4.04EE47 pKa = 3.63VKK49 pKa = 10.72IQVKK53 pKa = 10.5ASDD56 pKa = 4.11PNGDD60 pKa = 3.7SLVYY64 pKa = 9.04TWNATGGKK72 pKa = 9.57INGTGSEE79 pKa = 4.7VIWVAPQEE87 pKa = 4.05IGEE90 pKa = 4.06YY91 pKa = 9.22TITVKK96 pKa = 10.73VSDD99 pKa = 4.13SKK101 pKa = 11.77VDD103 pKa = 3.43VSKK106 pKa = 10.98SIQISVQPLPNEE118 pKa = 4.02APVISSLTVEE128 pKa = 4.35KK129 pKa = 10.86SVVQPEE135 pKa = 4.42EE136 pKa = 3.89KK137 pKa = 10.75VKK139 pKa = 11.03VQVQASDD146 pKa = 3.61PDD148 pKa = 4.39GDD150 pKa = 3.81VLTYY154 pKa = 9.15TWTVTGGSIEE164 pKa = 4.31GTGSEE169 pKa = 4.52VFWIASQTDD178 pKa = 3.44GNYY181 pKa = 9.57TITVEE186 pKa = 4.16VSDD189 pKa = 4.86GRR191 pKa = 11.84DD192 pKa = 3.09TVTGSINIQVNTAPIIEE209 pKa = 5.01EE210 pKa = 3.85ISASSSSVLVNGTLTLTAIVNDD232 pKa = 3.8PTGDD236 pKa = 3.31NLTYY240 pKa = 10.15RR241 pKa = 11.84WEE243 pKa = 4.13DD244 pKa = 3.41AIGEE248 pKa = 4.31VLGTEE253 pKa = 5.07PSLEE257 pKa = 3.6WHH259 pKa = 6.92APDD262 pKa = 3.5TYY264 pKa = 11.02GVYY267 pKa = 10.27PVTLYY272 pKa = 11.12VSDD275 pKa = 5.4GIFEE279 pKa = 4.14SSKK282 pKa = 9.61TININVEE289 pKa = 4.3DD290 pKa = 4.84DD291 pKa = 3.46SHH293 pKa = 6.5HH294 pKa = 6.41APEE297 pKa = 4.21IQEE300 pKa = 4.18VAITTPYY307 pKa = 10.45FEE309 pKa = 4.4SQVVVDD315 pKa = 4.53SVLAAGRR322 pKa = 11.84LYY324 pKa = 10.68AVWPIFVDD332 pKa = 3.44VDD334 pKa = 3.59EE335 pKa = 5.47SDD337 pKa = 3.53QLDD340 pKa = 3.63IQATADD346 pKa = 3.4VGEE349 pKa = 4.32VEE351 pKa = 4.66IFASEE356 pKa = 4.51YY357 pKa = 9.27EE358 pKa = 4.63TICFWVAPDD367 pKa = 3.4VAGEE371 pKa = 4.02YY372 pKa = 10.07TLTFTVTDD380 pKa = 4.77GIYY383 pKa = 9.78TVVKK387 pKa = 8.94EE388 pKa = 4.19YY389 pKa = 10.31PVQVVEE395 pKa = 4.32NSCPEE400 pKa = 3.31IDD402 pKa = 4.91AIYY405 pKa = 10.34INNEE409 pKa = 4.35LITDD413 pKa = 4.34NYY415 pKa = 8.63YY416 pKa = 10.15QAAPGEE422 pKa = 4.52EE423 pKa = 3.76ITISVLASDD432 pKa = 4.96SDD434 pKa = 3.6NDD436 pKa = 3.57APLEE440 pKa = 3.94YY441 pKa = 10.39KK442 pKa = 10.35IIDD445 pKa = 3.93PYY447 pKa = 10.62YY448 pKa = 10.04EE449 pKa = 3.42IWNTNEE455 pKa = 3.91LTFTAPMEE463 pKa = 4.16EE464 pKa = 4.34EE465 pKa = 3.86MDD467 pKa = 4.37EE468 pKa = 3.53IWIYY472 pKa = 11.28VFDD475 pKa = 4.21GLQVRR480 pKa = 11.84VASVIIDD487 pKa = 3.57VDD489 pKa = 3.48NGADD493 pKa = 4.44NNTNDD498 pKa = 5.13QNAIDD503 pKa = 4.35QNAIDD508 pKa = 4.57QINNLISRR516 pKa = 11.84FVNDD520 pKa = 5.01YY521 pKa = 10.62EE522 pKa = 4.27NQDD525 pKa = 3.25INSILSYY532 pKa = 10.6TDD534 pKa = 3.27SSYY537 pKa = 10.98QAEE540 pKa = 4.47LKK542 pKa = 10.68SRR544 pKa = 11.84LNRR547 pKa = 11.84YY548 pKa = 9.18FEE550 pKa = 4.51LEE552 pKa = 3.98RR553 pKa = 11.84VSNLTIRR560 pKa = 11.84KK561 pKa = 8.43LSEE564 pKa = 3.96IIEE567 pKa = 4.09VDD569 pKa = 3.47GEE571 pKa = 4.45FEE573 pKa = 4.19VQVAVQGDD581 pKa = 4.4FYY583 pKa = 11.72VLFEE587 pKa = 4.35TFPNSLIRR595 pKa = 11.84VFTFWFTYY603 pKa = 10.5DD604 pKa = 2.93EE605 pKa = 4.48YY606 pKa = 11.79GNILISSVWLL616 pKa = 3.62
Molecular weight: 68.34 kDa
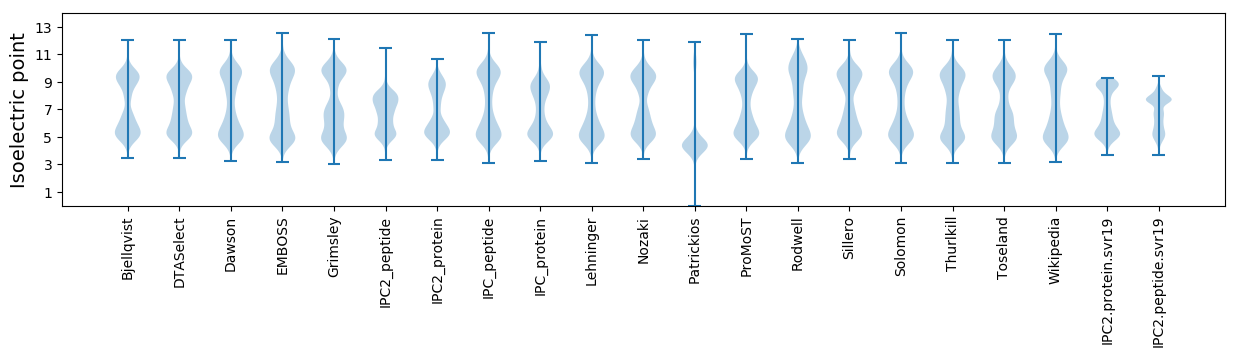
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S9SVD0|A0A3S9SVD0_9FIRM Na_H_Exchanger domain-containing protein OS=Anoxybacter fermentans OX=1323375 GN=BBF96_01770 PE=4 SV=1
MM1 pKa = 7.25NLRR4 pKa = 11.84YY5 pKa = 9.49RR6 pKa = 11.84RR7 pKa = 11.84NKK9 pKa = 10.5LMTRR13 pKa = 11.84CLINFIAILITGHH26 pKa = 6.91LLDD29 pKa = 5.39GIVVNSLIAVLAAAFVLGIVNTFIRR54 pKa = 11.84PILVILSLPLTIITLGLFTFVINALMLLLTASLIPGFSISSFWTAFFGAIIISILSSIILAVFDD118 pKa = 4.0
MM1 pKa = 7.25NLRR4 pKa = 11.84YY5 pKa = 9.49RR6 pKa = 11.84RR7 pKa = 11.84NKK9 pKa = 10.5LMTRR13 pKa = 11.84CLINFIAILITGHH26 pKa = 6.91LLDD29 pKa = 5.39GIVVNSLIAVLAAAFVLGIVNTFIRR54 pKa = 11.84PILVILSLPLTIITLGLFTFVINALMLLLTASLIPGFSISSFWTAFFGAIIISILSSIILAVFDD118 pKa = 4.0
Molecular weight: 12.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
927558 |
37 |
3256 |
308.1 |
34.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.174 ± 0.05 | 0.915 ± 0.017 |
5.097 ± 0.037 | 7.741 ± 0.05 |
4.403 ± 0.039 | 7.027 ± 0.048 |
1.624 ± 0.018 | 9.115 ± 0.038 |
7.76 ± 0.049 | 10.255 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.022 | 4.527 ± 0.037 |
3.642 ± 0.028 | 3.196 ± 0.029 |
4.682 ± 0.036 | 4.986 ± 0.032 |
4.708 ± 0.036 | 6.845 ± 0.042 |
0.963 ± 0.016 | 3.911 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
