
bacterium HR25
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
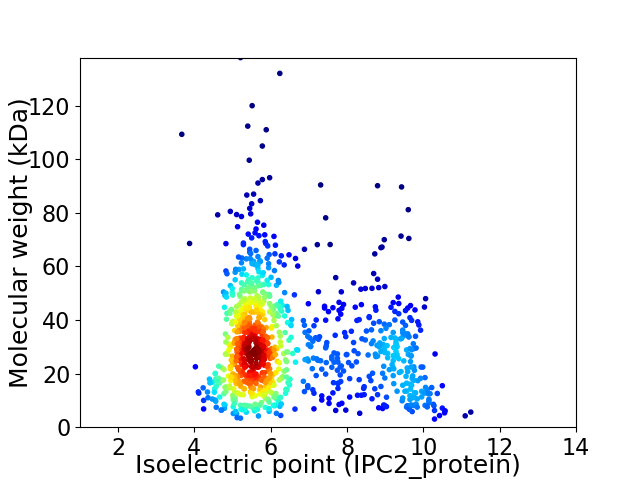
Virtual 2D-PAGE plot for 982 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5YL89|A0A2H5YL89_9BACT Acetyltransf_6 domain-containing protein OS=bacterium HR25 OX=2035420 GN=HRbin25_00039 PE=4 SV=1
MM1 pKa = 7.62LSGPARR7 pKa = 11.84RR8 pKa = 11.84PWGLLLAGVLLAVLTALAFGSDD30 pKa = 3.77SPALPGLGGPRR41 pKa = 11.84LAQAIPPHH49 pKa = 6.6SPALDD54 pKa = 3.61TDD56 pKa = 4.96DD57 pKa = 6.18DD58 pKa = 5.41DD59 pKa = 7.91DD60 pKa = 6.85DD61 pKa = 5.49ILVKK65 pKa = 10.84DD66 pKa = 4.18RR67 pKa = 11.84DD68 pKa = 3.74RR69 pKa = 11.84DD70 pKa = 3.96GVPDD74 pKa = 3.72STDD77 pKa = 3.09NCPRR81 pKa = 11.84TANASQEE88 pKa = 4.18DD89 pKa = 4.17TDD91 pKa = 4.21GDD93 pKa = 4.14GRR95 pKa = 11.84GDD97 pKa = 3.52ACDD100 pKa = 4.39PDD102 pKa = 4.44LDD104 pKa = 4.24NDD106 pKa = 4.32GVPNTADD113 pKa = 3.56KK114 pKa = 10.99RR115 pKa = 11.84PVPAGPGVRR124 pKa = 11.84ALDD127 pKa = 3.66ATTTDD132 pKa = 3.02SDD134 pKa = 4.09GDD136 pKa = 4.24GVPDD140 pKa = 5.02ASDD143 pKa = 3.2NCPFVPNASQEE154 pKa = 4.09DD155 pKa = 4.09TDD157 pKa = 4.62RR158 pKa = 11.84DD159 pKa = 4.48GIGDD163 pKa = 3.83ACDD166 pKa = 3.46TDD168 pKa = 3.68GDD170 pKa = 4.16NDD172 pKa = 4.21GVPDD176 pKa = 4.49SSDD179 pKa = 3.38NCPQDD184 pKa = 3.77PNPDD188 pKa = 3.58QVDD191 pKa = 3.36SDD193 pKa = 4.13GDD195 pKa = 4.22GEE197 pKa = 5.34GDD199 pKa = 3.28ACDD202 pKa = 3.51PVAEE206 pKa = 4.2VFYY209 pKa = 11.28VRR211 pKa = 11.84STDD214 pKa = 3.2GGQTFGNYY222 pKa = 9.13YY223 pKa = 10.49NISNDD228 pKa = 3.35GLHH231 pKa = 6.67SGIAHH236 pKa = 6.05VAASGGVVHH245 pKa = 6.76IVWDD249 pKa = 3.98RR250 pKa = 11.84GDD252 pKa = 3.38EE253 pKa = 3.98PRR255 pKa = 11.84EE256 pKa = 3.71VVYY259 pKa = 10.57RR260 pKa = 11.84RR261 pKa = 11.84SLDD264 pKa = 3.45GGVSFAPIFNASSNNGDD281 pKa = 3.44SSEE284 pKa = 4.34SDD286 pKa = 3.3LTVGGSVLHH295 pKa = 5.89VVWEE299 pKa = 4.07EE300 pKa = 4.21DD301 pKa = 3.63VLDD304 pKa = 4.6ANGNPDD310 pKa = 3.73PCCPGDD316 pKa = 3.24EE317 pKa = 4.11AYY319 pKa = 10.67YY320 pKa = 10.39RR321 pKa = 11.84RR322 pKa = 11.84SGDD325 pKa = 3.13AGQTFSASRR334 pKa = 11.84NLSQTPNAHH343 pKa = 6.53SRR345 pKa = 11.84DD346 pKa = 3.71PDD348 pKa = 3.57VATSGNLVGVVHH360 pKa = 7.14EE361 pKa = 5.59DD362 pKa = 3.45PTPGNDD368 pKa = 3.69DD369 pKa = 3.38IFFEE373 pKa = 4.53RR374 pKa = 11.84STDD377 pKa = 4.25GGLTWSTFVNLSQDD391 pKa = 3.48AGQSAEE397 pKa = 3.97PAIDD401 pKa = 3.14ISGNTVHH408 pKa = 6.36VVWEE412 pKa = 4.29HH413 pKa = 5.84RR414 pKa = 11.84ASGRR418 pKa = 11.84IAYY421 pKa = 8.68VRR423 pKa = 11.84STDD426 pKa = 4.7GGVTWSAPLLLPAGADD442 pKa = 3.17RR443 pKa = 11.84AAIVAASGSTVHH455 pKa = 6.49VVACGGGEE463 pKa = 3.74VRR465 pKa = 11.84YY466 pKa = 10.09YY467 pKa = 10.97RR468 pKa = 11.84STDD471 pKa = 3.15GGATWGAPVNLSNNPGACSKK491 pKa = 10.95PMVDD495 pKa = 3.3VLGDD499 pKa = 3.23QVYY502 pKa = 8.83VVWEE506 pKa = 4.19DD507 pKa = 3.65TTTGASDD514 pKa = 3.53VYY516 pKa = 10.87LRR518 pKa = 11.84RR519 pKa = 11.84SSDD522 pKa = 2.99GGITFAPTQNLSNNAGEE539 pKa = 4.19SSEE542 pKa = 4.15PSVAVDD548 pKa = 3.5PSTGHH553 pKa = 6.53VYY555 pKa = 9.85VVWYY559 pKa = 10.0DD560 pKa = 3.89FTRR563 pKa = 11.84PTPPAAPADD572 pKa = 4.0ADD574 pKa = 3.84SDD576 pKa = 4.34GVADD580 pKa = 5.28PSDD583 pKa = 3.82NCPGVANPDD592 pKa = 3.32QADD595 pKa = 3.47ADD597 pKa = 4.14GDD599 pKa = 4.36GQGDD603 pKa = 3.73ACDD606 pKa = 4.69PDD608 pKa = 4.99DD609 pKa = 6.42DD610 pKa = 5.06NDD612 pKa = 4.21GVPDD616 pKa = 4.56ASDD619 pKa = 3.46NCPFAANAAQTDD631 pKa = 3.73TDD633 pKa = 4.04GDD635 pKa = 4.34GQGDD639 pKa = 3.91ACDD642 pKa = 4.48SDD644 pKa = 5.23DD645 pKa = 5.93DD646 pKa = 5.14GDD648 pKa = 4.27GVADD652 pKa = 4.56PFDD655 pKa = 4.14NCPLVPNAGQEE666 pKa = 4.12NTDD669 pKa = 3.35GDD671 pKa = 4.51GQGDD675 pKa = 3.93ACDD678 pKa = 5.58DD679 pKa = 4.59DD680 pKa = 6.45DD681 pKa = 7.02DD682 pKa = 6.22NDD684 pKa = 5.29GIPDD688 pKa = 3.9ASDD691 pKa = 3.66PCPLQSDD698 pKa = 4.32CDD700 pKa = 4.05SDD702 pKa = 4.48GVPDD706 pKa = 4.17SSDD709 pKa = 3.19NCRR712 pKa = 11.84LAFNPAQQDD721 pKa = 3.16SDD723 pKa = 3.84GDD725 pKa = 4.23GTGDD729 pKa = 4.36ACDD732 pKa = 3.6TDD734 pKa = 4.61GNPMLAFVAAGPGGATLYY752 pKa = 10.6EE753 pKa = 4.76FGRR756 pKa = 11.84GTTGEE761 pKa = 3.3IWYY764 pKa = 9.21RR765 pKa = 11.84AWDD768 pKa = 3.78GTAWSAWTGLSAVANSPPTVATSGADD794 pKa = 3.05LYY796 pKa = 11.63VFARR800 pKa = 11.84GSDD803 pKa = 3.28DD804 pKa = 3.5GLWYY808 pKa = 10.35RR809 pKa = 11.84RR810 pKa = 11.84LSGGTWDD817 pKa = 3.09AWTPLGGTMRR827 pKa = 11.84GVPAAVATAGGHH839 pKa = 5.4IYY841 pKa = 10.8VFVRR845 pKa = 11.84GASGDD850 pKa = 2.37IWYY853 pKa = 9.52RR854 pKa = 11.84HH855 pKa = 5.22WDD857 pKa = 3.42GTAWQPWASLGGLATEE873 pKa = 4.72SPGALALDD881 pKa = 3.71SEE883 pKa = 4.94VFLFVRR889 pKa = 11.84GSDD892 pKa = 3.23DD893 pKa = 3.49GLWYY897 pKa = 10.33RR898 pKa = 11.84RR899 pKa = 11.84LSGATWGDD907 pKa = 3.42WASLGGPVSSGPRR920 pKa = 11.84PVALGSVEE928 pKa = 4.15ADD930 pKa = 3.16VFARR934 pKa = 11.84GTDD937 pKa = 3.17NAVWYY942 pKa = 8.67RR943 pKa = 11.84HH944 pKa = 5.27WDD946 pKa = 3.43GTTWGAWQTLGGSLHH961 pKa = 6.79SPPQPLAVGGSLFLMALGTGDD982 pKa = 4.23DD983 pKa = 3.17LWYY986 pKa = 10.16RR987 pKa = 11.84QLSGGTWADD996 pKa = 3.32WQPLEE1001 pKa = 4.45GRR1003 pKa = 11.84LTAAPALVAFQSGAFVFGRR1022 pKa = 11.84GSEE1025 pKa = 3.97GDD1027 pKa = 2.84VWYY1030 pKa = 10.41RR1031 pKa = 11.84HH1032 pKa = 5.49WDD1034 pKa = 3.41GTTWGAWQPLGGLVTAGVPP1053 pKa = 3.42
MM1 pKa = 7.62LSGPARR7 pKa = 11.84RR8 pKa = 11.84PWGLLLAGVLLAVLTALAFGSDD30 pKa = 3.77SPALPGLGGPRR41 pKa = 11.84LAQAIPPHH49 pKa = 6.6SPALDD54 pKa = 3.61TDD56 pKa = 4.96DD57 pKa = 6.18DD58 pKa = 5.41DD59 pKa = 7.91DD60 pKa = 6.85DD61 pKa = 5.49ILVKK65 pKa = 10.84DD66 pKa = 4.18RR67 pKa = 11.84DD68 pKa = 3.74RR69 pKa = 11.84DD70 pKa = 3.96GVPDD74 pKa = 3.72STDD77 pKa = 3.09NCPRR81 pKa = 11.84TANASQEE88 pKa = 4.18DD89 pKa = 4.17TDD91 pKa = 4.21GDD93 pKa = 4.14GRR95 pKa = 11.84GDD97 pKa = 3.52ACDD100 pKa = 4.39PDD102 pKa = 4.44LDD104 pKa = 4.24NDD106 pKa = 4.32GVPNTADD113 pKa = 3.56KK114 pKa = 10.99RR115 pKa = 11.84PVPAGPGVRR124 pKa = 11.84ALDD127 pKa = 3.66ATTTDD132 pKa = 3.02SDD134 pKa = 4.09GDD136 pKa = 4.24GVPDD140 pKa = 5.02ASDD143 pKa = 3.2NCPFVPNASQEE154 pKa = 4.09DD155 pKa = 4.09TDD157 pKa = 4.62RR158 pKa = 11.84DD159 pKa = 4.48GIGDD163 pKa = 3.83ACDD166 pKa = 3.46TDD168 pKa = 3.68GDD170 pKa = 4.16NDD172 pKa = 4.21GVPDD176 pKa = 4.49SSDD179 pKa = 3.38NCPQDD184 pKa = 3.77PNPDD188 pKa = 3.58QVDD191 pKa = 3.36SDD193 pKa = 4.13GDD195 pKa = 4.22GEE197 pKa = 5.34GDD199 pKa = 3.28ACDD202 pKa = 3.51PVAEE206 pKa = 4.2VFYY209 pKa = 11.28VRR211 pKa = 11.84STDD214 pKa = 3.2GGQTFGNYY222 pKa = 9.13YY223 pKa = 10.49NISNDD228 pKa = 3.35GLHH231 pKa = 6.67SGIAHH236 pKa = 6.05VAASGGVVHH245 pKa = 6.76IVWDD249 pKa = 3.98RR250 pKa = 11.84GDD252 pKa = 3.38EE253 pKa = 3.98PRR255 pKa = 11.84EE256 pKa = 3.71VVYY259 pKa = 10.57RR260 pKa = 11.84RR261 pKa = 11.84SLDD264 pKa = 3.45GGVSFAPIFNASSNNGDD281 pKa = 3.44SSEE284 pKa = 4.34SDD286 pKa = 3.3LTVGGSVLHH295 pKa = 5.89VVWEE299 pKa = 4.07EE300 pKa = 4.21DD301 pKa = 3.63VLDD304 pKa = 4.6ANGNPDD310 pKa = 3.73PCCPGDD316 pKa = 3.24EE317 pKa = 4.11AYY319 pKa = 10.67YY320 pKa = 10.39RR321 pKa = 11.84RR322 pKa = 11.84SGDD325 pKa = 3.13AGQTFSASRR334 pKa = 11.84NLSQTPNAHH343 pKa = 6.53SRR345 pKa = 11.84DD346 pKa = 3.71PDD348 pKa = 3.57VATSGNLVGVVHH360 pKa = 7.14EE361 pKa = 5.59DD362 pKa = 3.45PTPGNDD368 pKa = 3.69DD369 pKa = 3.38IFFEE373 pKa = 4.53RR374 pKa = 11.84STDD377 pKa = 4.25GGLTWSTFVNLSQDD391 pKa = 3.48AGQSAEE397 pKa = 3.97PAIDD401 pKa = 3.14ISGNTVHH408 pKa = 6.36VVWEE412 pKa = 4.29HH413 pKa = 5.84RR414 pKa = 11.84ASGRR418 pKa = 11.84IAYY421 pKa = 8.68VRR423 pKa = 11.84STDD426 pKa = 4.7GGVTWSAPLLLPAGADD442 pKa = 3.17RR443 pKa = 11.84AAIVAASGSTVHH455 pKa = 6.49VVACGGGEE463 pKa = 3.74VRR465 pKa = 11.84YY466 pKa = 10.09YY467 pKa = 10.97RR468 pKa = 11.84STDD471 pKa = 3.15GGATWGAPVNLSNNPGACSKK491 pKa = 10.95PMVDD495 pKa = 3.3VLGDD499 pKa = 3.23QVYY502 pKa = 8.83VVWEE506 pKa = 4.19DD507 pKa = 3.65TTTGASDD514 pKa = 3.53VYY516 pKa = 10.87LRR518 pKa = 11.84RR519 pKa = 11.84SSDD522 pKa = 2.99GGITFAPTQNLSNNAGEE539 pKa = 4.19SSEE542 pKa = 4.15PSVAVDD548 pKa = 3.5PSTGHH553 pKa = 6.53VYY555 pKa = 9.85VVWYY559 pKa = 10.0DD560 pKa = 3.89FTRR563 pKa = 11.84PTPPAAPADD572 pKa = 4.0ADD574 pKa = 3.84SDD576 pKa = 4.34GVADD580 pKa = 5.28PSDD583 pKa = 3.82NCPGVANPDD592 pKa = 3.32QADD595 pKa = 3.47ADD597 pKa = 4.14GDD599 pKa = 4.36GQGDD603 pKa = 3.73ACDD606 pKa = 4.69PDD608 pKa = 4.99DD609 pKa = 6.42DD610 pKa = 5.06NDD612 pKa = 4.21GVPDD616 pKa = 4.56ASDD619 pKa = 3.46NCPFAANAAQTDD631 pKa = 3.73TDD633 pKa = 4.04GDD635 pKa = 4.34GQGDD639 pKa = 3.91ACDD642 pKa = 4.48SDD644 pKa = 5.23DD645 pKa = 5.93DD646 pKa = 5.14GDD648 pKa = 4.27GVADD652 pKa = 4.56PFDD655 pKa = 4.14NCPLVPNAGQEE666 pKa = 4.12NTDD669 pKa = 3.35GDD671 pKa = 4.51GQGDD675 pKa = 3.93ACDD678 pKa = 5.58DD679 pKa = 4.59DD680 pKa = 6.45DD681 pKa = 7.02DD682 pKa = 6.22NDD684 pKa = 5.29GIPDD688 pKa = 3.9ASDD691 pKa = 3.66PCPLQSDD698 pKa = 4.32CDD700 pKa = 4.05SDD702 pKa = 4.48GVPDD706 pKa = 4.17SSDD709 pKa = 3.19NCRR712 pKa = 11.84LAFNPAQQDD721 pKa = 3.16SDD723 pKa = 3.84GDD725 pKa = 4.23GTGDD729 pKa = 4.36ACDD732 pKa = 3.6TDD734 pKa = 4.61GNPMLAFVAAGPGGATLYY752 pKa = 10.6EE753 pKa = 4.76FGRR756 pKa = 11.84GTTGEE761 pKa = 3.3IWYY764 pKa = 9.21RR765 pKa = 11.84AWDD768 pKa = 3.78GTAWSAWTGLSAVANSPPTVATSGADD794 pKa = 3.05LYY796 pKa = 11.63VFARR800 pKa = 11.84GSDD803 pKa = 3.28DD804 pKa = 3.5GLWYY808 pKa = 10.35RR809 pKa = 11.84RR810 pKa = 11.84LSGGTWDD817 pKa = 3.09AWTPLGGTMRR827 pKa = 11.84GVPAAVATAGGHH839 pKa = 5.4IYY841 pKa = 10.8VFVRR845 pKa = 11.84GASGDD850 pKa = 2.37IWYY853 pKa = 9.52RR854 pKa = 11.84HH855 pKa = 5.22WDD857 pKa = 3.42GTAWQPWASLGGLATEE873 pKa = 4.72SPGALALDD881 pKa = 3.71SEE883 pKa = 4.94VFLFVRR889 pKa = 11.84GSDD892 pKa = 3.23DD893 pKa = 3.49GLWYY897 pKa = 10.33RR898 pKa = 11.84RR899 pKa = 11.84LSGATWGDD907 pKa = 3.42WASLGGPVSSGPRR920 pKa = 11.84PVALGSVEE928 pKa = 4.15ADD930 pKa = 3.16VFARR934 pKa = 11.84GTDD937 pKa = 3.17NAVWYY942 pKa = 8.67RR943 pKa = 11.84HH944 pKa = 5.27WDD946 pKa = 3.43GTTWGAWQTLGGSLHH961 pKa = 6.79SPPQPLAVGGSLFLMALGTGDD982 pKa = 4.23DD983 pKa = 3.17LWYY986 pKa = 10.16RR987 pKa = 11.84QLSGGTWADD996 pKa = 3.32WQPLEE1001 pKa = 4.45GRR1003 pKa = 11.84LTAAPALVAFQSGAFVFGRR1022 pKa = 11.84GSEE1025 pKa = 3.97GDD1027 pKa = 2.84VWYY1030 pKa = 10.41RR1031 pKa = 11.84HH1032 pKa = 5.49WDD1034 pKa = 3.41GTTWGAWQPLGGLVTAGVPP1053 pKa = 3.42
Molecular weight: 109.34 kDa
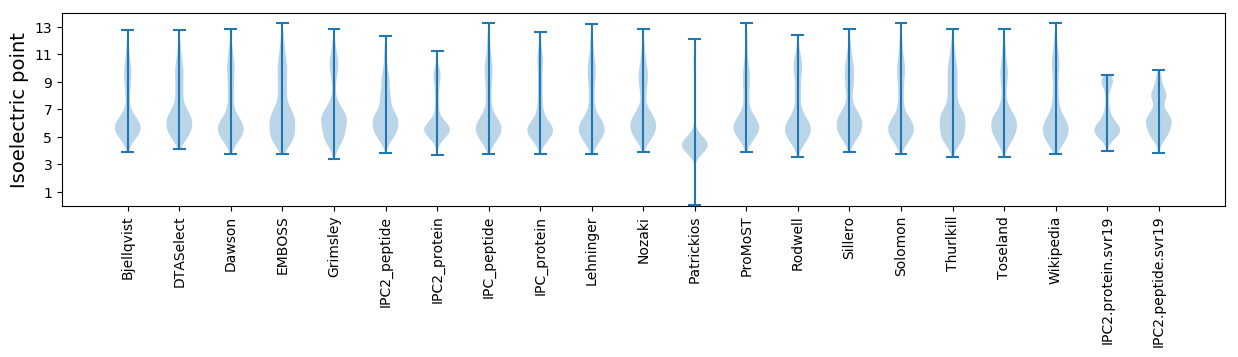
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5YMZ8|A0A2H5YMZ8_9BACT Acetolactate synthase large subunit IlvG OS=bacterium HR25 OX=2035420 GN=ilvG_2 PE=3 SV=1
MM1 pKa = 8.03PKK3 pKa = 8.99RR4 pKa = 11.84TYY6 pKa = 9.41QPKK9 pKa = 8.73NRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84QRR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 5.04GFLARR22 pKa = 11.84NRR24 pKa = 11.84TRR26 pKa = 11.84SGRR29 pKa = 11.84AVLKK33 pKa = 10.34RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.82GRR40 pKa = 11.84WRR42 pKa = 11.84LAVV45 pKa = 3.35
MM1 pKa = 8.03PKK3 pKa = 8.99RR4 pKa = 11.84TYY6 pKa = 9.41QPKK9 pKa = 8.73NRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84QRR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 5.04GFLARR22 pKa = 11.84NRR24 pKa = 11.84TRR26 pKa = 11.84SGRR29 pKa = 11.84AVLKK33 pKa = 10.34RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.82GRR40 pKa = 11.84WRR42 pKa = 11.84LAVV45 pKa = 3.35
Molecular weight: 5.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
285183 |
30 |
1256 |
290.4 |
31.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.366 ± 0.098 | 0.935 ± 0.026 |
4.508 ± 0.059 | 7.503 ± 0.082 |
3.046 ± 0.047 | 9.062 ± 0.08 |
2.064 ± 0.039 | 3.865 ± 0.067 |
1.83 ± 0.046 | 11.839 ± 0.101 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.032 | 1.729 ± 0.036 |
6.154 ± 0.066 | 3.624 ± 0.054 |
9.238 ± 0.084 | 4.399 ± 0.05 |
4.317 ± 0.049 | 8.456 ± 0.063 |
1.482 ± 0.04 | 2.578 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
