
Candidatus Synechococcus spongiarum LMB bulk10E
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; Candidatus Synechococcus spongiarum
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
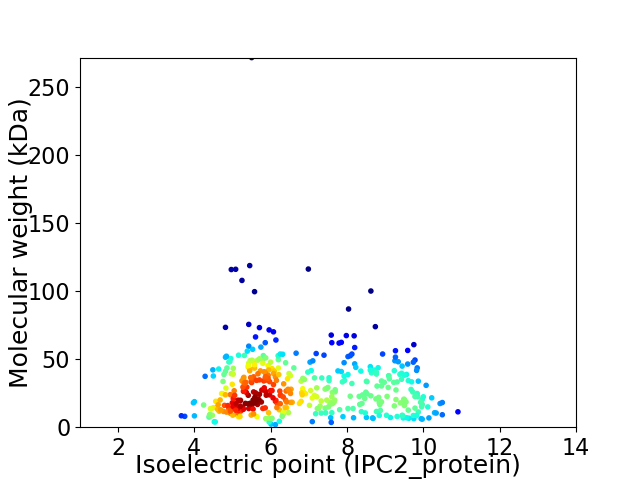
Virtual 2D-PAGE plot for 427 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N4ARF1|A0A6N4ARF1_9SYNE Uncharacterized protein OS=Candidatus Synechococcus spongiarum LMB bulk10E OX=1449122 GN=BO91_00140 PE=4 SV=1
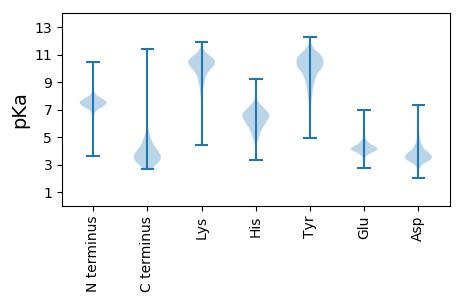
MM1 pKa = 6.84YY2 pKa = 10.22QNKK5 pKa = 9.65SLSHH9 pKa = 6.81NYY11 pKa = 8.18TAITNHH17 pKa = 6.29CLDD20 pKa = 3.02TGMYY24 pKa = 8.87FGCIKK29 pKa = 10.33GFPGAHH35 pKa = 5.56SQGEE39 pKa = 4.41TLDD42 pKa = 3.77EE43 pKa = 4.98LNTNLQEE50 pKa = 4.44VISMLNDD57 pKa = 3.33DD58 pKa = 4.77NVPVFIDD65 pKa = 3.13NNEE68 pKa = 4.01YY69 pKa = 10.7VDD71 pKa = 3.8RR72 pKa = 11.84QTFISII78 pKa = 3.85
MM1 pKa = 6.84YY2 pKa = 10.22QNKK5 pKa = 9.65SLSHH9 pKa = 6.81NYY11 pKa = 8.18TAITNHH17 pKa = 6.29CLDD20 pKa = 3.02TGMYY24 pKa = 8.87FGCIKK29 pKa = 10.33GFPGAHH35 pKa = 5.56SQGEE39 pKa = 4.41TLDD42 pKa = 3.77EE43 pKa = 4.98LNTNLQEE50 pKa = 4.44VISMLNDD57 pKa = 3.33DD58 pKa = 4.77NVPVFIDD65 pKa = 3.13NNEE68 pKa = 4.01YY69 pKa = 10.7VDD71 pKa = 3.8RR72 pKa = 11.84QTFISII78 pKa = 3.85
Molecular weight: 8.85 kDa
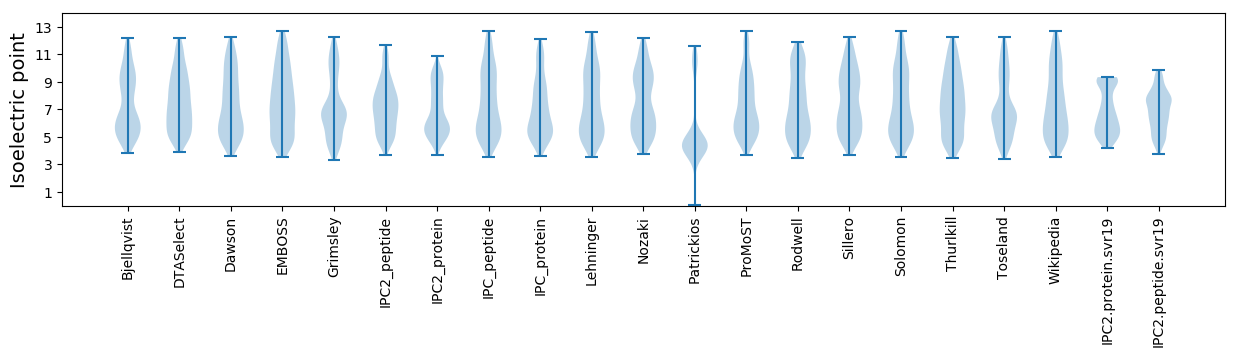
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N4APX0|A0A6N4APX0_9SYNE Uncharacterized protein OS=Candidatus Synechococcus spongiarum LMB bulk10E OX=1449122 GN=BO91_02025 PE=4 SV=1
MM1 pKa = 7.65ALLNGSPLGQRR12 pKa = 11.84PTGIGVVTLALAKK25 pKa = 9.97ALCHH29 pKa = 5.83EE30 pKa = 4.78RR31 pKa = 11.84VTFLDD36 pKa = 3.91PVATGRR42 pKa = 11.84PNSLAIPGNLSMDD55 pKa = 3.94HH56 pKa = 5.92GRR58 pKa = 11.84KK59 pKa = 8.71GHH61 pKa = 6.01LRR63 pKa = 11.84RR64 pKa = 11.84LLWTQNQLPGLLKK77 pKa = 9.96KK78 pKa = 9.26TGTSLLLSPLPEE90 pKa = 5.54APMLRR95 pKa = 11.84RR96 pKa = 11.84GVRR99 pKa = 11.84SIVLAHH105 pKa = 6.74DD106 pKa = 5.56LIPLRR111 pKa = 11.84FPRR114 pKa = 11.84PGPLLAYY121 pKa = 9.75HH122 pKa = 7.06LAYY125 pKa = 10.52VPLVLHH131 pKa = 4.88QAEE134 pKa = 3.87RR135 pKa = 11.84VLCNSEE141 pKa = 3.43ATARR145 pKa = 11.84EE146 pKa = 3.58IHH148 pKa = 5.93HH149 pKa = 6.73WLRR152 pKa = 11.84VPLRR156 pKa = 11.84KK157 pKa = 9.97LITVPLGFDD166 pKa = 3.64PGQLRR171 pKa = 11.84PLDD174 pKa = 3.86VPRR177 pKa = 11.84QPFFLVLGRR186 pKa = 11.84HH187 pKa = 6.66DD188 pKa = 3.63YY189 pKa = 10.92HH190 pKa = 7.67KK191 pKa = 10.7NLPRR195 pKa = 11.84ILRR198 pKa = 11.84AFARR202 pKa = 11.84IADD205 pKa = 3.84PDD207 pKa = 3.74MTLKK211 pKa = 11.01LVGSPHH217 pKa = 7.22AYY219 pKa = 8.44LTAKK223 pKa = 10.2LKK225 pKa = 10.75HH226 pKa = 5.35LAARR230 pKa = 11.84LRR232 pKa = 11.84IAHH235 pKa = 4.98QCQWIHH241 pKa = 5.15WVSDD245 pKa = 3.48AEE247 pKa = 4.45RR248 pKa = 11.84LRR250 pKa = 11.84LLNTAKK256 pKa = 10.77ALVMPSLWEE265 pKa = 4.25GFGLPALEE273 pKa = 5.22AMACGTPVIAGNAGALSEE291 pKa = 4.48VVGSVGLLVNPTNTSHH307 pKa = 7.18IAEE310 pKa = 4.39AMADD314 pKa = 3.55VLSDD318 pKa = 3.94NKK320 pKa = 10.23IQRR323 pKa = 11.84QARR326 pKa = 11.84LGGPSRR332 pKa = 11.84AARR335 pKa = 11.84FRR337 pKa = 11.84WDD339 pKa = 2.88NTARR343 pKa = 11.84QVEE346 pKa = 4.46SLLEE350 pKa = 4.04NFF352 pKa = 4.8
MM1 pKa = 7.65ALLNGSPLGQRR12 pKa = 11.84PTGIGVVTLALAKK25 pKa = 9.97ALCHH29 pKa = 5.83EE30 pKa = 4.78RR31 pKa = 11.84VTFLDD36 pKa = 3.91PVATGRR42 pKa = 11.84PNSLAIPGNLSMDD55 pKa = 3.94HH56 pKa = 5.92GRR58 pKa = 11.84KK59 pKa = 8.71GHH61 pKa = 6.01LRR63 pKa = 11.84RR64 pKa = 11.84LLWTQNQLPGLLKK77 pKa = 9.96KK78 pKa = 9.26TGTSLLLSPLPEE90 pKa = 5.54APMLRR95 pKa = 11.84RR96 pKa = 11.84GVRR99 pKa = 11.84SIVLAHH105 pKa = 6.74DD106 pKa = 5.56LIPLRR111 pKa = 11.84FPRR114 pKa = 11.84PGPLLAYY121 pKa = 9.75HH122 pKa = 7.06LAYY125 pKa = 10.52VPLVLHH131 pKa = 4.88QAEE134 pKa = 3.87RR135 pKa = 11.84VLCNSEE141 pKa = 3.43ATARR145 pKa = 11.84EE146 pKa = 3.58IHH148 pKa = 5.93HH149 pKa = 6.73WLRR152 pKa = 11.84VPLRR156 pKa = 11.84KK157 pKa = 9.97LITVPLGFDD166 pKa = 3.64PGQLRR171 pKa = 11.84PLDD174 pKa = 3.86VPRR177 pKa = 11.84QPFFLVLGRR186 pKa = 11.84HH187 pKa = 6.66DD188 pKa = 3.63YY189 pKa = 10.92HH190 pKa = 7.67KK191 pKa = 10.7NLPRR195 pKa = 11.84ILRR198 pKa = 11.84AFARR202 pKa = 11.84IADD205 pKa = 3.84PDD207 pKa = 3.74MTLKK211 pKa = 11.01LVGSPHH217 pKa = 7.22AYY219 pKa = 8.44LTAKK223 pKa = 10.2LKK225 pKa = 10.75HH226 pKa = 5.35LAARR230 pKa = 11.84LRR232 pKa = 11.84IAHH235 pKa = 4.98QCQWIHH241 pKa = 5.15WVSDD245 pKa = 3.48AEE247 pKa = 4.45RR248 pKa = 11.84LRR250 pKa = 11.84LLNTAKK256 pKa = 10.77ALVMPSLWEE265 pKa = 4.25GFGLPALEE273 pKa = 5.22AMACGTPVIAGNAGALSEE291 pKa = 4.48VVGSVGLLVNPTNTSHH307 pKa = 7.18IAEE310 pKa = 4.39AMADD314 pKa = 3.55VLSDD318 pKa = 3.94NKK320 pKa = 10.23IQRR323 pKa = 11.84QARR326 pKa = 11.84LGGPSRR332 pKa = 11.84AARR335 pKa = 11.84FRR337 pKa = 11.84WDD339 pKa = 2.88NTARR343 pKa = 11.84QVEE346 pKa = 4.46SLLEE350 pKa = 4.04NFF352 pKa = 4.8
Molecular weight: 38.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
116829 |
17 |
2453 |
273.6 |
30.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.147 ± 0.172 | 1.399 ± 0.048 |
5.094 ± 0.089 | 5.452 ± 0.131 |
3.117 ± 0.079 | 8.232 ± 0.114 |
2.54 ± 0.079 | 3.955 ± 0.102 |
2.747 ± 0.125 | 11.964 ± 0.167 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.1 ± 0.054 | 2.628 ± 0.074 |
5.791 ± 0.102 | 4.779 ± 0.091 |
7.663 ± 0.119 | 5.903 ± 0.109 |
5.387 ± 0.125 | 7.451 ± 0.108 |
1.631 ± 0.055 | 2.022 ± 0.063 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
