
Human picobirnavirus (strain Human/Thailand/Hy005102/-) (PBV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Picobirnaviridae; Picobirnavirus; Human picobirnavirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
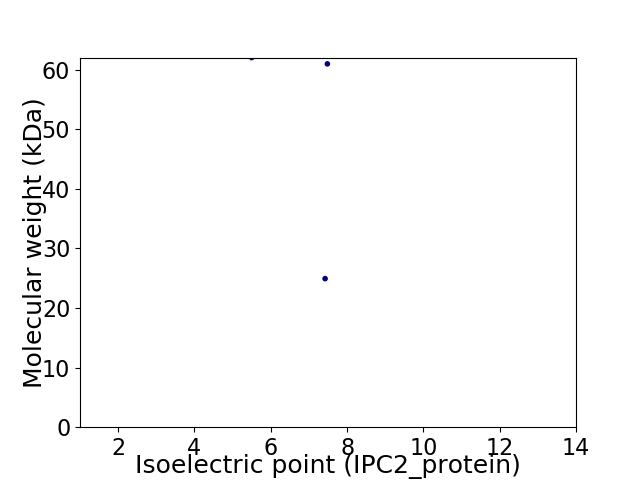
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
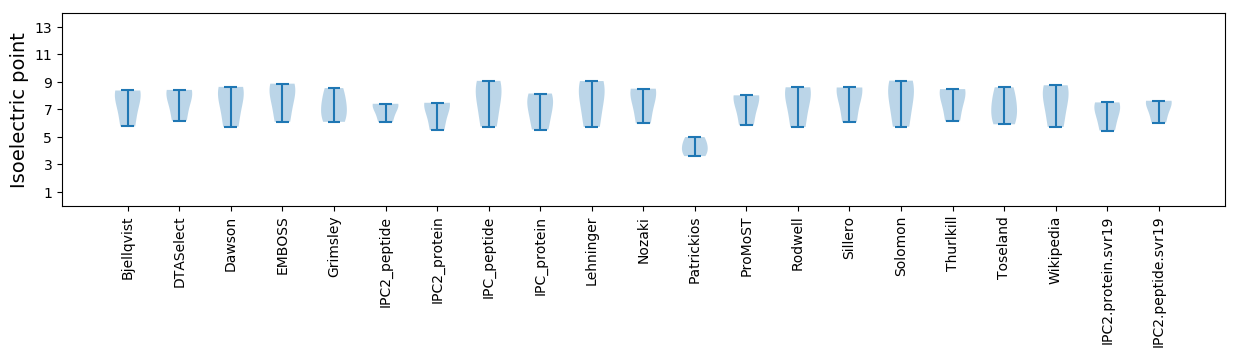
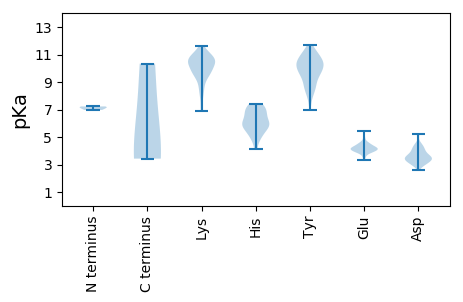
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q50LE6|ORF1_HPBVH Uncharacterized ORF1 protein OS=Human picobirnavirus (strain Human/Thailand/Hy005102/-) OX=647332 GN=Segment-1 PE=4 SV=1
MM1 pKa = 7.14KK2 pKa = 10.7QNDD5 pKa = 4.09TKK7 pKa = 10.72KK8 pKa = 6.93TTQRR12 pKa = 11.84RR13 pKa = 11.84NSKK16 pKa = 10.14KK17 pKa = 10.38YY18 pKa = 9.4SSKK21 pKa = 9.73TNRR24 pKa = 11.84GTKK27 pKa = 9.57RR28 pKa = 11.84APRR31 pKa = 11.84DD32 pKa = 3.52QEE34 pKa = 4.18VGTGAQEE41 pKa = 3.97STRR44 pKa = 11.84NDD46 pKa = 3.4VAWYY50 pKa = 10.23ARR52 pKa = 11.84YY53 pKa = 8.65PHH55 pKa = 7.38ILEE58 pKa = 4.27EE59 pKa = 4.17ATRR62 pKa = 11.84LPFAYY67 pKa = 9.51PIGQYY72 pKa = 10.67YY73 pKa = 7.98DD74 pKa = 2.97TGYY77 pKa = 10.75SVASATEE84 pKa = 3.58WSKK87 pKa = 11.59YY88 pKa = 9.77VDD90 pKa = 3.43TSLTIPGVMCVNFTPTPGEE109 pKa = 4.32SYY111 pKa = 11.1NKK113 pKa = 10.04NSPINIAAQNVYY125 pKa = 10.08TYY127 pKa = 10.27VRR129 pKa = 11.84HH130 pKa = 5.77MNSGHH135 pKa = 6.72ANYY138 pKa = 9.57EE139 pKa = 4.01QADD142 pKa = 3.63LMMYY146 pKa = 10.3LLAMDD151 pKa = 4.35SLYY154 pKa = 10.49IFHH157 pKa = 7.1SYY159 pKa = 7.64VRR161 pKa = 11.84KK162 pKa = 9.72ILAISKK168 pKa = 10.38LYY170 pKa = 10.45TPVNKK175 pKa = 10.07YY176 pKa = 9.55FPRR179 pKa = 11.84ALLVALGVDD188 pKa = 3.84PEE190 pKa = 4.92DD191 pKa = 3.58VFANQAQWEE200 pKa = 4.65YY201 pKa = 9.74FVNMVAYY208 pKa = 10.26RR209 pKa = 11.84AGAFAAPASMTYY221 pKa = 8.88YY222 pKa = 10.18EE223 pKa = 3.98RR224 pKa = 11.84HH225 pKa = 4.89AWMSNGLYY233 pKa = 10.02VDD235 pKa = 3.52QDD237 pKa = 3.46VTRR240 pKa = 11.84AQIYY244 pKa = 6.99MFKK247 pKa = 9.65PTMLWKK253 pKa = 10.67YY254 pKa = 10.02EE255 pKa = 4.01NLGTTGTKK263 pKa = 9.96LVPLMMPKK271 pKa = 10.51AGDD274 pKa = 3.34NRR276 pKa = 11.84KK277 pKa = 9.33LVDD280 pKa = 4.11FQVLFNNLVSTMLGDD295 pKa = 3.57EE296 pKa = 5.0DD297 pKa = 4.39FGIMSGDD304 pKa = 3.37VFKK307 pKa = 11.38AFGADD312 pKa = 3.34GLVKK316 pKa = 10.53LLAVDD321 pKa = 4.02STTMTLPTYY330 pKa = 10.66DD331 pKa = 4.53PLILAQIHH339 pKa = 5.3SARR342 pKa = 11.84AVGAPILEE350 pKa = 4.42TSTLTGFPGRR360 pKa = 11.84QWQITQNPDD369 pKa = 2.85VNNGAIIFHH378 pKa = 7.01PSFGYY383 pKa = 10.41DD384 pKa = 3.4GQDD387 pKa = 3.14HH388 pKa = 6.95EE389 pKa = 4.76EE390 pKa = 3.86LSFRR394 pKa = 11.84AMCSNMILNLPGEE407 pKa = 4.19AHH409 pKa = 5.82SAEE412 pKa = 4.51MIIEE416 pKa = 4.25ATRR419 pKa = 11.84LATMFQVKK427 pKa = 9.63AVPAGDD433 pKa = 3.35TSKK436 pKa = 10.89PVLYY440 pKa = 10.23LPNGFGTEE448 pKa = 4.11VVNDD452 pKa = 3.48YY453 pKa = 11.01TMISVDD459 pKa = 3.15KK460 pKa = 9.82ATPHH464 pKa = 7.31DD465 pKa = 4.23LTIHH469 pKa = 5.79TFFNNILVPNAKK481 pKa = 9.74EE482 pKa = 3.85NYY484 pKa = 8.24VANLEE489 pKa = 4.14LLNNIIQFDD498 pKa = 4.25WAPQLYY504 pKa = 8.16LTYY507 pKa = 10.72GIAQEE512 pKa = 4.37SFGPFAQLNDD522 pKa = 3.39WTILTGEE529 pKa = 4.27TLARR533 pKa = 11.84MHH535 pKa = 6.16EE536 pKa = 4.32VCVTSMFDD544 pKa = 3.43VPQMGFNKK552 pKa = 10.34
MM1 pKa = 7.14KK2 pKa = 10.7QNDD5 pKa = 4.09TKK7 pKa = 10.72KK8 pKa = 6.93TTQRR12 pKa = 11.84RR13 pKa = 11.84NSKK16 pKa = 10.14KK17 pKa = 10.38YY18 pKa = 9.4SSKK21 pKa = 9.73TNRR24 pKa = 11.84GTKK27 pKa = 9.57RR28 pKa = 11.84APRR31 pKa = 11.84DD32 pKa = 3.52QEE34 pKa = 4.18VGTGAQEE41 pKa = 3.97STRR44 pKa = 11.84NDD46 pKa = 3.4VAWYY50 pKa = 10.23ARR52 pKa = 11.84YY53 pKa = 8.65PHH55 pKa = 7.38ILEE58 pKa = 4.27EE59 pKa = 4.17ATRR62 pKa = 11.84LPFAYY67 pKa = 9.51PIGQYY72 pKa = 10.67YY73 pKa = 7.98DD74 pKa = 2.97TGYY77 pKa = 10.75SVASATEE84 pKa = 3.58WSKK87 pKa = 11.59YY88 pKa = 9.77VDD90 pKa = 3.43TSLTIPGVMCVNFTPTPGEE109 pKa = 4.32SYY111 pKa = 11.1NKK113 pKa = 10.04NSPINIAAQNVYY125 pKa = 10.08TYY127 pKa = 10.27VRR129 pKa = 11.84HH130 pKa = 5.77MNSGHH135 pKa = 6.72ANYY138 pKa = 9.57EE139 pKa = 4.01QADD142 pKa = 3.63LMMYY146 pKa = 10.3LLAMDD151 pKa = 4.35SLYY154 pKa = 10.49IFHH157 pKa = 7.1SYY159 pKa = 7.64VRR161 pKa = 11.84KK162 pKa = 9.72ILAISKK168 pKa = 10.38LYY170 pKa = 10.45TPVNKK175 pKa = 10.07YY176 pKa = 9.55FPRR179 pKa = 11.84ALLVALGVDD188 pKa = 3.84PEE190 pKa = 4.92DD191 pKa = 3.58VFANQAQWEE200 pKa = 4.65YY201 pKa = 9.74FVNMVAYY208 pKa = 10.26RR209 pKa = 11.84AGAFAAPASMTYY221 pKa = 8.88YY222 pKa = 10.18EE223 pKa = 3.98RR224 pKa = 11.84HH225 pKa = 4.89AWMSNGLYY233 pKa = 10.02VDD235 pKa = 3.52QDD237 pKa = 3.46VTRR240 pKa = 11.84AQIYY244 pKa = 6.99MFKK247 pKa = 9.65PTMLWKK253 pKa = 10.67YY254 pKa = 10.02EE255 pKa = 4.01NLGTTGTKK263 pKa = 9.96LVPLMMPKK271 pKa = 10.51AGDD274 pKa = 3.34NRR276 pKa = 11.84KK277 pKa = 9.33LVDD280 pKa = 4.11FQVLFNNLVSTMLGDD295 pKa = 3.57EE296 pKa = 5.0DD297 pKa = 4.39FGIMSGDD304 pKa = 3.37VFKK307 pKa = 11.38AFGADD312 pKa = 3.34GLVKK316 pKa = 10.53LLAVDD321 pKa = 4.02STTMTLPTYY330 pKa = 10.66DD331 pKa = 4.53PLILAQIHH339 pKa = 5.3SARR342 pKa = 11.84AVGAPILEE350 pKa = 4.42TSTLTGFPGRR360 pKa = 11.84QWQITQNPDD369 pKa = 2.85VNNGAIIFHH378 pKa = 7.01PSFGYY383 pKa = 10.41DD384 pKa = 3.4GQDD387 pKa = 3.14HH388 pKa = 6.95EE389 pKa = 4.76EE390 pKa = 3.86LSFRR394 pKa = 11.84AMCSNMILNLPGEE407 pKa = 4.19AHH409 pKa = 5.82SAEE412 pKa = 4.51MIIEE416 pKa = 4.25ATRR419 pKa = 11.84LATMFQVKK427 pKa = 9.63AVPAGDD433 pKa = 3.35TSKK436 pKa = 10.89PVLYY440 pKa = 10.23LPNGFGTEE448 pKa = 4.11VVNDD452 pKa = 3.48YY453 pKa = 11.01TMISVDD459 pKa = 3.15KK460 pKa = 9.82ATPHH464 pKa = 7.31DD465 pKa = 4.23LTIHH469 pKa = 5.79TFFNNILVPNAKK481 pKa = 9.74EE482 pKa = 3.85NYY484 pKa = 8.24VANLEE489 pKa = 4.14LLNNIIQFDD498 pKa = 4.25WAPQLYY504 pKa = 8.16LTYY507 pKa = 10.72GIAQEE512 pKa = 4.37SFGPFAQLNDD522 pKa = 3.39WTILTGEE529 pKa = 4.27TLARR533 pKa = 11.84MHH535 pKa = 6.16EE536 pKa = 4.32VCVTSMFDD544 pKa = 3.43VPQMGFNKK552 pKa = 10.34
Molecular weight: 62.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q50LE6|ORF1_HPBVH Uncharacterized ORF1 protein OS=Human picobirnavirus (strain Human/Thailand/Hy005102/-) OX=647332 GN=Segment-1 PE=4 SV=1
MM1 pKa = 7.23TANQIAYY8 pKa = 9.41QKK10 pKa = 9.95HH11 pKa = 6.14LEE13 pKa = 4.19TARR16 pKa = 11.84VNAVGEE22 pKa = 4.17MQRR25 pKa = 11.84GLEE28 pKa = 3.89LDD30 pKa = 3.21EE31 pKa = 4.5SRR33 pKa = 11.84RR34 pKa = 11.84HH35 pKa = 6.31NISQEE40 pKa = 3.84QLKK43 pKa = 8.74TRR45 pKa = 11.84EE46 pKa = 4.1LTEE49 pKa = 4.06LEE51 pKa = 4.02RR52 pKa = 11.84SNRR55 pKa = 11.84AVEE58 pKa = 4.37KK59 pKa = 8.64EE60 pKa = 4.25TSRR63 pKa = 11.84HH64 pKa = 4.11NVVTEE69 pKa = 3.92TEE71 pKa = 4.18TRR73 pKa = 11.84RR74 pKa = 11.84SNLARR79 pKa = 11.84EE80 pKa = 4.2WEE82 pKa = 4.58TYY84 pKa = 9.78RR85 pKa = 11.84SNSARR90 pKa = 11.84EE91 pKa = 4.02MEE93 pKa = 4.39TQRR96 pKa = 11.84SNISYY101 pKa = 9.5EE102 pKa = 4.08AIKK105 pKa = 10.39RR106 pKa = 11.84GQLALDD112 pKa = 3.36RR113 pKa = 11.84AEE115 pKa = 4.19LNEE118 pKa = 4.57SIRR121 pKa = 11.84ATNEE125 pKa = 3.33NLALQYY131 pKa = 11.24SKK133 pKa = 11.22LQTEE137 pKa = 4.27SLLTQRR143 pKa = 11.84GQDD146 pKa = 3.68LQHH149 pKa = 6.65KK150 pKa = 8.89NAIIGASANAFGSLLGYY167 pKa = 8.59STASADD173 pKa = 3.62RR174 pKa = 11.84ASRR177 pKa = 11.84EE178 pKa = 4.2EE179 pKa = 3.74IASANRR185 pKa = 11.84KK186 pKa = 7.17SQEE189 pKa = 4.54HH190 pKa = 5.82IASMQVLGSMANTMFSSVSNLVGKK214 pKa = 7.87TAGAFAGGLSS224 pKa = 3.43
MM1 pKa = 7.23TANQIAYY8 pKa = 9.41QKK10 pKa = 9.95HH11 pKa = 6.14LEE13 pKa = 4.19TARR16 pKa = 11.84VNAVGEE22 pKa = 4.17MQRR25 pKa = 11.84GLEE28 pKa = 3.89LDD30 pKa = 3.21EE31 pKa = 4.5SRR33 pKa = 11.84RR34 pKa = 11.84HH35 pKa = 6.31NISQEE40 pKa = 3.84QLKK43 pKa = 8.74TRR45 pKa = 11.84EE46 pKa = 4.1LTEE49 pKa = 4.06LEE51 pKa = 4.02RR52 pKa = 11.84SNRR55 pKa = 11.84AVEE58 pKa = 4.37KK59 pKa = 8.64EE60 pKa = 4.25TSRR63 pKa = 11.84HH64 pKa = 4.11NVVTEE69 pKa = 3.92TEE71 pKa = 4.18TRR73 pKa = 11.84RR74 pKa = 11.84SNLARR79 pKa = 11.84EE80 pKa = 4.2WEE82 pKa = 4.58TYY84 pKa = 9.78RR85 pKa = 11.84SNSARR90 pKa = 11.84EE91 pKa = 4.02MEE93 pKa = 4.39TQRR96 pKa = 11.84SNISYY101 pKa = 9.5EE102 pKa = 4.08AIKK105 pKa = 10.39RR106 pKa = 11.84GQLALDD112 pKa = 3.36RR113 pKa = 11.84AEE115 pKa = 4.19LNEE118 pKa = 4.57SIRR121 pKa = 11.84ATNEE125 pKa = 3.33NLALQYY131 pKa = 11.24SKK133 pKa = 11.22LQTEE137 pKa = 4.27SLLTQRR143 pKa = 11.84GQDD146 pKa = 3.68LQHH149 pKa = 6.65KK150 pKa = 8.89NAIIGASANAFGSLLGYY167 pKa = 8.59STASADD173 pKa = 3.62RR174 pKa = 11.84ASRR177 pKa = 11.84EE178 pKa = 4.2EE179 pKa = 3.74IASANRR185 pKa = 11.84KK186 pKa = 7.17SQEE189 pKa = 4.54HH190 pKa = 5.82IASMQVLGSMANTMFSSVSNLVGKK214 pKa = 7.87TAGAFAGGLSS224 pKa = 3.43
Molecular weight: 24.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1310 |
224 |
552 |
436.7 |
49.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.015 ± 1.279 | 0.916 ± 0.385 |
5.267 ± 1.024 | 5.725 ± 1.457 |
3.588 ± 0.781 | 6.412 ± 0.446 |
2.061 ± 0.092 | 4.427 ± 0.172 |
4.962 ± 0.606 | 8.244 ± 0.321 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.893 ± 0.378 | 5.267 ± 0.928 |
4.275 ± 1.246 | 4.504 ± 0.495 |
5.725 ± 1.366 | 6.87 ± 1.338 |
6.489 ± 0.914 | 6.87 ± 0.999 |
1.756 ± 0.464 | 4.733 ± 0.766 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
