
Rhizobium sp. NFR07
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
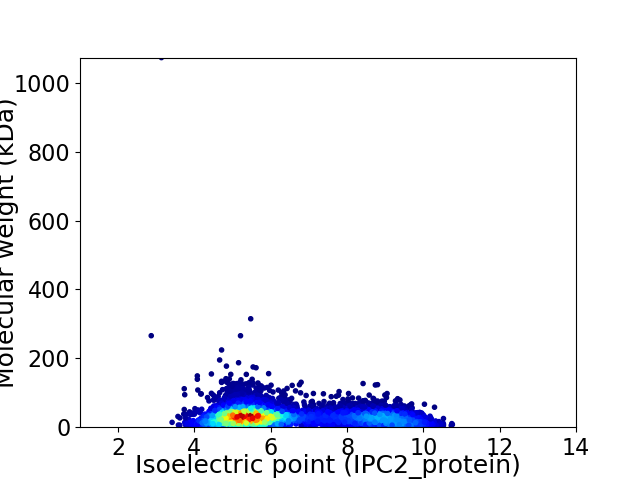
Virtual 2D-PAGE plot for 6556 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1CAW4|A0A1I1CAW4_9RHIZ Uncharacterized protein OS=Rhizobium sp. NFR07 OX=1566262 GN=SAMN03159496_05495 PE=4 SV=1
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAAVSGAQAADD24 pKa = 4.21AIVAAEE30 pKa = 4.47PEE32 pKa = 3.93PMEE35 pKa = 4.1YY36 pKa = 10.8VRR38 pKa = 11.84VCDD41 pKa = 4.42AFGTGYY47 pKa = 10.47FYY49 pKa = 10.57IPGTEE54 pKa = 3.81TCLKK58 pKa = 9.41VGGRR62 pKa = 11.84VRR64 pKa = 11.84FDD66 pKa = 2.39AWYY69 pKa = 10.61GDD71 pKa = 3.72VYY73 pKa = 11.45DD74 pKa = 5.16NEE76 pKa = 5.74DD77 pKa = 3.34GTFTNTRR84 pKa = 11.84AEE86 pKa = 4.17IYY88 pKa = 10.33LDD90 pKa = 3.67SASDD94 pKa = 4.05TEE96 pKa = 4.31WGALKK101 pKa = 10.01TSIVARR107 pKa = 11.84FDD109 pKa = 4.25YY110 pKa = 10.86NPHH113 pKa = 6.56NDD115 pKa = 4.24DD116 pKa = 3.93GNGGAFNDD124 pKa = 3.76ATRR127 pKa = 11.84TRR129 pKa = 11.84LIGANIQLGGFLVGLADD146 pKa = 3.81SYY148 pKa = 11.66YY149 pKa = 11.18SSFINYY155 pKa = 10.03AGDD158 pKa = 3.74VINDD162 pKa = 3.73DD163 pKa = 4.22VISYY167 pKa = 10.93GPFEE171 pKa = 5.21LNQVAYY177 pKa = 10.84VFDD180 pKa = 4.61AGNGFGAYY188 pKa = 9.06IAVEE192 pKa = 4.16DD193 pKa = 5.19DD194 pKa = 3.69GQNDD198 pKa = 4.02DD199 pKa = 5.88DD200 pKa = 4.76IPGPASDD207 pKa = 4.09WVDD210 pKa = 3.29VTAGVKK216 pKa = 9.94YY217 pKa = 11.02DD218 pKa = 3.82NGTFMAGLVGGYY230 pKa = 9.84DD231 pKa = 3.43EE232 pKa = 5.18SVEE235 pKa = 4.04EE236 pKa = 4.28GAIKK240 pKa = 10.53ARR242 pKa = 11.84IGGTFGAFSAFILGGWNTDD261 pKa = 2.44GDD263 pKa = 4.57TLNKK267 pKa = 9.49YY268 pKa = 10.56APGDD272 pKa = 3.66SLGNPWGDD280 pKa = 2.9WALWAGAGYY289 pKa = 10.53KK290 pKa = 10.51FSDD293 pKa = 3.95KK294 pKa = 10.43IAANVQGSVTDD305 pKa = 4.06FGTWAITGNVQWTPVSGLLIQPEE328 pKa = 4.41LSYY331 pKa = 11.29TDD333 pKa = 3.57WEE335 pKa = 4.78DD336 pKa = 4.23GGDD339 pKa = 3.56QFGGLIRR346 pKa = 11.84IQRR349 pKa = 11.84TFF351 pKa = 2.87
MM1 pKa = 7.49NIKK4 pKa = 10.39SLLLGSAAALAAVSGAQAADD24 pKa = 4.21AIVAAEE30 pKa = 4.47PEE32 pKa = 3.93PMEE35 pKa = 4.1YY36 pKa = 10.8VRR38 pKa = 11.84VCDD41 pKa = 4.42AFGTGYY47 pKa = 10.47FYY49 pKa = 10.57IPGTEE54 pKa = 3.81TCLKK58 pKa = 9.41VGGRR62 pKa = 11.84VRR64 pKa = 11.84FDD66 pKa = 2.39AWYY69 pKa = 10.61GDD71 pKa = 3.72VYY73 pKa = 11.45DD74 pKa = 5.16NEE76 pKa = 5.74DD77 pKa = 3.34GTFTNTRR84 pKa = 11.84AEE86 pKa = 4.17IYY88 pKa = 10.33LDD90 pKa = 3.67SASDD94 pKa = 4.05TEE96 pKa = 4.31WGALKK101 pKa = 10.01TSIVARR107 pKa = 11.84FDD109 pKa = 4.25YY110 pKa = 10.86NPHH113 pKa = 6.56NDD115 pKa = 4.24DD116 pKa = 3.93GNGGAFNDD124 pKa = 3.76ATRR127 pKa = 11.84TRR129 pKa = 11.84LIGANIQLGGFLVGLADD146 pKa = 3.81SYY148 pKa = 11.66YY149 pKa = 11.18SSFINYY155 pKa = 10.03AGDD158 pKa = 3.74VINDD162 pKa = 3.73DD163 pKa = 4.22VISYY167 pKa = 10.93GPFEE171 pKa = 5.21LNQVAYY177 pKa = 10.84VFDD180 pKa = 4.61AGNGFGAYY188 pKa = 9.06IAVEE192 pKa = 4.16DD193 pKa = 5.19DD194 pKa = 3.69GQNDD198 pKa = 4.02DD199 pKa = 5.88DD200 pKa = 4.76IPGPASDD207 pKa = 4.09WVDD210 pKa = 3.29VTAGVKK216 pKa = 9.94YY217 pKa = 11.02DD218 pKa = 3.82NGTFMAGLVGGYY230 pKa = 9.84DD231 pKa = 3.43EE232 pKa = 5.18SVEE235 pKa = 4.04EE236 pKa = 4.28GAIKK240 pKa = 10.53ARR242 pKa = 11.84IGGTFGAFSAFILGGWNTDD261 pKa = 2.44GDD263 pKa = 4.57TLNKK267 pKa = 9.49YY268 pKa = 10.56APGDD272 pKa = 3.66SLGNPWGDD280 pKa = 2.9WALWAGAGYY289 pKa = 10.53KK290 pKa = 10.51FSDD293 pKa = 3.95KK294 pKa = 10.43IAANVQGSVTDD305 pKa = 4.06FGTWAITGNVQWTPVSGLLIQPEE328 pKa = 4.41LSYY331 pKa = 11.29TDD333 pKa = 3.57WEE335 pKa = 4.78DD336 pKa = 4.23GGDD339 pKa = 3.56QFGGLIRR346 pKa = 11.84IQRR349 pKa = 11.84TFF351 pKa = 2.87
Molecular weight: 37.41 kDa
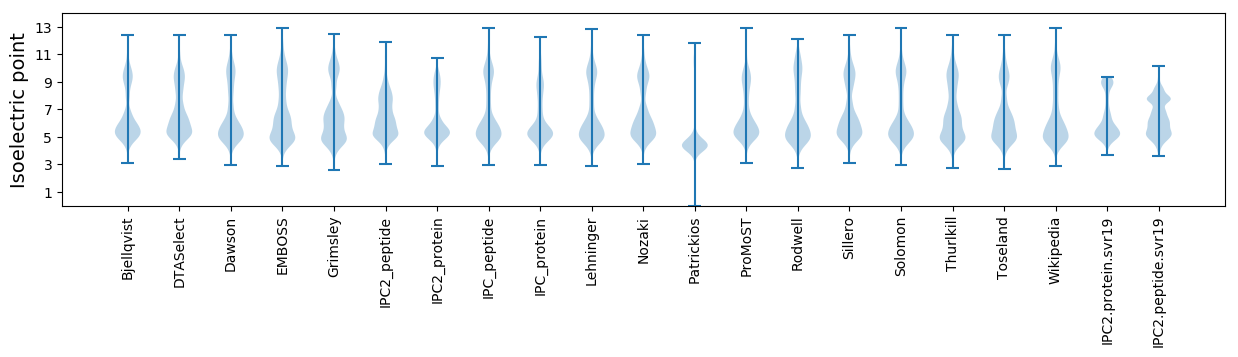
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1CUQ2|A0A1I1CUQ2_9RHIZ DNA-binding transcriptional regulator LysR family OS=Rhizobium sp. NFR07 OX=1566262 GN=SAMN03159496_06403 PE=3 SV=1
MM1 pKa = 7.15TEE3 pKa = 3.95IVAGSIDD10 pKa = 3.73PSGTRR15 pKa = 11.84ASSPASHH22 pKa = 6.57TEE24 pKa = 3.8YY25 pKa = 10.75VKK27 pKa = 10.42GWRR30 pKa = 11.84IAAICIVLLLINAFSQIDD48 pKa = 3.92RR49 pKa = 11.84ILPFILAEE57 pKa = 4.45KK58 pKa = 10.26IKK60 pKa = 10.98ADD62 pKa = 4.33LSLTDD67 pKa = 3.51TQMGLLTGIAFAVCYY82 pKa = 10.32ALLSLPLARR91 pKa = 11.84AADD94 pKa = 3.91RR95 pKa = 11.84GSPRR99 pKa = 11.84LVLVSCILLWSVMTTLGGFAASFMFLALTRR129 pKa = 11.84FGVAFGEE136 pKa = 4.5AGAVPAGHH144 pKa = 6.93ALIVRR149 pKa = 11.84KK150 pKa = 9.6IGHH153 pKa = 5.83EE154 pKa = 3.87RR155 pKa = 11.84RR156 pKa = 11.84GLAIGLFAMGIPLGTMFGFAAGGAIADD183 pKa = 3.52IFGWRR188 pKa = 11.84IVLMGAGAVGILIGLLTILIAGPTPALQSKK218 pKa = 9.95AGRR221 pKa = 11.84SEE223 pKa = 3.88PFLRR227 pKa = 11.84TSLEE231 pKa = 4.18MLSSPAFRR239 pKa = 11.84WMFTGAIFLGFAGAPFYY256 pKa = 11.17AFSIPFLIRR265 pKa = 11.84THH267 pKa = 6.51GLTATEE273 pKa = 4.34VGVSFGMLQGLMGIIGTLGGGRR295 pKa = 11.84WFDD298 pKa = 3.2HH299 pKa = 6.33AVRR302 pKa = 11.84SGTGRR307 pKa = 11.84VLGPPAILFLIASVTTTAALFAPAGWMSVALFVPAMLSFSFFLPWGFGAAHH358 pKa = 6.86LVAGPGRR365 pKa = 11.84QAQATSLMMIGSGLLGPALAPLFVGIVSDD394 pKa = 3.8AASAADD400 pKa = 3.53IPNGLGLGMLIVPVAAVSTGITLLIANRR428 pKa = 11.84RR429 pKa = 11.84IGAFLRR435 pKa = 11.84QPP437 pKa = 3.37
MM1 pKa = 7.15TEE3 pKa = 3.95IVAGSIDD10 pKa = 3.73PSGTRR15 pKa = 11.84ASSPASHH22 pKa = 6.57TEE24 pKa = 3.8YY25 pKa = 10.75VKK27 pKa = 10.42GWRR30 pKa = 11.84IAAICIVLLLINAFSQIDD48 pKa = 3.92RR49 pKa = 11.84ILPFILAEE57 pKa = 4.45KK58 pKa = 10.26IKK60 pKa = 10.98ADD62 pKa = 4.33LSLTDD67 pKa = 3.51TQMGLLTGIAFAVCYY82 pKa = 10.32ALLSLPLARR91 pKa = 11.84AADD94 pKa = 3.91RR95 pKa = 11.84GSPRR99 pKa = 11.84LVLVSCILLWSVMTTLGGFAASFMFLALTRR129 pKa = 11.84FGVAFGEE136 pKa = 4.5AGAVPAGHH144 pKa = 6.93ALIVRR149 pKa = 11.84KK150 pKa = 9.6IGHH153 pKa = 5.83EE154 pKa = 3.87RR155 pKa = 11.84RR156 pKa = 11.84GLAIGLFAMGIPLGTMFGFAAGGAIADD183 pKa = 3.52IFGWRR188 pKa = 11.84IVLMGAGAVGILIGLLTILIAGPTPALQSKK218 pKa = 9.95AGRR221 pKa = 11.84SEE223 pKa = 3.88PFLRR227 pKa = 11.84TSLEE231 pKa = 4.18MLSSPAFRR239 pKa = 11.84WMFTGAIFLGFAGAPFYY256 pKa = 11.17AFSIPFLIRR265 pKa = 11.84THH267 pKa = 6.51GLTATEE273 pKa = 4.34VGVSFGMLQGLMGIIGTLGGGRR295 pKa = 11.84WFDD298 pKa = 3.2HH299 pKa = 6.33AVRR302 pKa = 11.84SGTGRR307 pKa = 11.84VLGPPAILFLIASVTTTAALFAPAGWMSVALFVPAMLSFSFFLPWGFGAAHH358 pKa = 6.86LVAGPGRR365 pKa = 11.84QAQATSLMMIGSGLLGPALAPLFVGIVSDD394 pKa = 3.8AASAADD400 pKa = 3.53IPNGLGLGMLIVPVAAVSTGITLLIANRR428 pKa = 11.84RR429 pKa = 11.84IGAFLRR435 pKa = 11.84QPP437 pKa = 3.37
Molecular weight: 45.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1986648 |
21 |
10674 |
303.0 |
32.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.766 ± 0.041 | 0.77 ± 0.008 |
5.784 ± 0.031 | 5.814 ± 0.03 |
3.902 ± 0.022 | 8.348 ± 0.029 |
1.984 ± 0.015 | 5.75 ± 0.019 |
3.657 ± 0.028 | 9.834 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.652 ± 0.016 | 2.825 ± 0.024 |
4.888 ± 0.02 | 3.148 ± 0.019 |
6.595 ± 0.038 | 5.926 ± 0.022 |
5.441 ± 0.051 | 7.326 ± 0.027 |
1.281 ± 0.013 | 2.309 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
