
Xylanimonas cellulosilytica (strain DSM 15894 / CECT 5975 / LMG 20990 / XIL07)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Promicromonosporaceae; Xylanimonas; Xylanimonas cellulosilytica
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
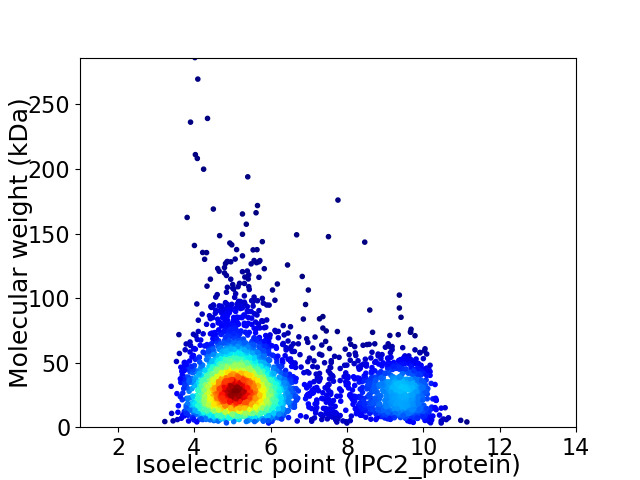
Virtual 2D-PAGE plot for 3441 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D1C0I5|D1C0I5_XYLCX Uncharacterized protein OS=Xylanimonas cellulosilytica (strain DSM 15894 / CECT 5975 / LMG 20990 / XIL07) OX=446471 GN=Xcel_3188 PE=4 SV=1
MM1 pKa = 7.45LGVGDD6 pKa = 5.12ADD8 pKa = 3.76PTKK11 pKa = 9.52EE12 pKa = 3.95PQMTRR17 pKa = 11.84STMRR21 pKa = 11.84PTARR25 pKa = 11.84LSARR29 pKa = 11.84PAARR33 pKa = 11.84LAAAAATATCLVAVAACSGPSDD55 pKa = 4.01AGTSASDD62 pKa = 3.71DD63 pKa = 3.62ATVGPLEE70 pKa = 4.21AMITEE75 pKa = 5.01IVGDD79 pKa = 3.93WNSAEE84 pKa = 4.06QQAKK88 pKa = 8.36QMEE91 pKa = 4.48MEE93 pKa = 4.19EE94 pKa = 4.02AVARR98 pKa = 11.84CMADD102 pKa = 2.64EE103 pKa = 4.5GFEE106 pKa = 4.09YY107 pKa = 10.88TPVDD111 pKa = 3.78YY112 pKa = 10.65SALMAEE118 pKa = 4.51VDD120 pKa = 4.09AVDD123 pKa = 3.51TTTRR127 pKa = 11.84EE128 pKa = 3.86YY129 pKa = 10.65AQEE132 pKa = 3.78YY133 pKa = 9.94GYY135 pKa = 11.12GHH137 pKa = 7.3SIQPDD142 pKa = 3.63TTGNPMPEE150 pKa = 3.52FVDD153 pKa = 3.74PNEE156 pKa = 4.8DD157 pKa = 3.18YY158 pKa = 10.97VAAMSEE164 pKa = 4.55SEE166 pKa = 4.12SEE168 pKa = 4.22AYY170 pKa = 10.57YY171 pKa = 10.22EE172 pKa = 4.06ALYY175 pKa = 10.81GAMEE179 pKa = 4.31YY180 pKa = 10.76DD181 pKa = 3.58VDD183 pKa = 4.16NVDD186 pKa = 5.28SIPDD190 pKa = 3.69MSEE193 pKa = 3.67MGCTGAAQVEE203 pKa = 4.55IMGQQDD209 pKa = 3.03TSLFDD214 pKa = 3.93NEE216 pKa = 4.02EE217 pKa = 3.77LTAFGEE223 pKa = 4.2NMEE226 pKa = 4.12QLEE229 pKa = 4.58ADD231 pKa = 3.71VAADD235 pKa = 3.66PKK237 pKa = 10.65VAEE240 pKa = 4.75AAAGWAEE247 pKa = 4.06CMADD251 pKa = 3.82AGFDD255 pKa = 3.49YY256 pKa = 8.61ATPQEE261 pKa = 4.29AQDD264 pKa = 4.3DD265 pKa = 4.45FMTRR269 pKa = 11.84SQAIWEE275 pKa = 4.26EE276 pKa = 3.94ADD278 pKa = 3.88PEE280 pKa = 4.56SVDD283 pKa = 5.36APEE286 pKa = 4.24QDD288 pKa = 4.28ADD290 pKa = 3.77LQEE293 pKa = 4.32QEE295 pKa = 4.65RR296 pKa = 11.84EE297 pKa = 3.92AAVADD302 pKa = 4.57FDD304 pKa = 4.65CKK306 pKa = 10.99EE307 pKa = 4.0KK308 pKa = 10.99VGYY311 pKa = 9.71DD312 pKa = 3.74AIRR315 pKa = 11.84RR316 pKa = 11.84EE317 pKa = 4.04VQFALEE323 pKa = 3.77QEE325 pKa = 4.67YY326 pKa = 10.41IDD328 pKa = 3.5THH330 pKa = 5.01EE331 pKa = 5.05AEE333 pKa = 4.49LEE335 pKa = 3.99AVKK338 pKa = 10.43AAFEE342 pKa = 4.38SLGLL346 pKa = 3.82
MM1 pKa = 7.45LGVGDD6 pKa = 5.12ADD8 pKa = 3.76PTKK11 pKa = 9.52EE12 pKa = 3.95PQMTRR17 pKa = 11.84STMRR21 pKa = 11.84PTARR25 pKa = 11.84LSARR29 pKa = 11.84PAARR33 pKa = 11.84LAAAAATATCLVAVAACSGPSDD55 pKa = 4.01AGTSASDD62 pKa = 3.71DD63 pKa = 3.62ATVGPLEE70 pKa = 4.21AMITEE75 pKa = 5.01IVGDD79 pKa = 3.93WNSAEE84 pKa = 4.06QQAKK88 pKa = 8.36QMEE91 pKa = 4.48MEE93 pKa = 4.19EE94 pKa = 4.02AVARR98 pKa = 11.84CMADD102 pKa = 2.64EE103 pKa = 4.5GFEE106 pKa = 4.09YY107 pKa = 10.88TPVDD111 pKa = 3.78YY112 pKa = 10.65SALMAEE118 pKa = 4.51VDD120 pKa = 4.09AVDD123 pKa = 3.51TTTRR127 pKa = 11.84EE128 pKa = 3.86YY129 pKa = 10.65AQEE132 pKa = 3.78YY133 pKa = 9.94GYY135 pKa = 11.12GHH137 pKa = 7.3SIQPDD142 pKa = 3.63TTGNPMPEE150 pKa = 3.52FVDD153 pKa = 3.74PNEE156 pKa = 4.8DD157 pKa = 3.18YY158 pKa = 10.97VAAMSEE164 pKa = 4.55SEE166 pKa = 4.12SEE168 pKa = 4.22AYY170 pKa = 10.57YY171 pKa = 10.22EE172 pKa = 4.06ALYY175 pKa = 10.81GAMEE179 pKa = 4.31YY180 pKa = 10.76DD181 pKa = 3.58VDD183 pKa = 4.16NVDD186 pKa = 5.28SIPDD190 pKa = 3.69MSEE193 pKa = 3.67MGCTGAAQVEE203 pKa = 4.55IMGQQDD209 pKa = 3.03TSLFDD214 pKa = 3.93NEE216 pKa = 4.02EE217 pKa = 3.77LTAFGEE223 pKa = 4.2NMEE226 pKa = 4.12QLEE229 pKa = 4.58ADD231 pKa = 3.71VAADD235 pKa = 3.66PKK237 pKa = 10.65VAEE240 pKa = 4.75AAAGWAEE247 pKa = 4.06CMADD251 pKa = 3.82AGFDD255 pKa = 3.49YY256 pKa = 8.61ATPQEE261 pKa = 4.29AQDD264 pKa = 4.3DD265 pKa = 4.45FMTRR269 pKa = 11.84SQAIWEE275 pKa = 4.26EE276 pKa = 3.94ADD278 pKa = 3.88PEE280 pKa = 4.56SVDD283 pKa = 5.36APEE286 pKa = 4.24QDD288 pKa = 4.28ADD290 pKa = 3.77LQEE293 pKa = 4.32QEE295 pKa = 4.65RR296 pKa = 11.84EE297 pKa = 3.92AAVADD302 pKa = 4.57FDD304 pKa = 4.65CKK306 pKa = 10.99EE307 pKa = 4.0KK308 pKa = 10.99VGYY311 pKa = 9.71DD312 pKa = 3.74AIRR315 pKa = 11.84RR316 pKa = 11.84EE317 pKa = 4.04VQFALEE323 pKa = 3.77QEE325 pKa = 4.67YY326 pKa = 10.41IDD328 pKa = 3.5THH330 pKa = 5.01EE331 pKa = 5.05AEE333 pKa = 4.49LEE335 pKa = 3.99AVKK338 pKa = 10.43AAFEE342 pKa = 4.38SLGLL346 pKa = 3.82
Molecular weight: 37.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D1BW16|D1BW16_XYLCX Proton-translocating NADH-quinone oxidoreductase chain L OS=Xylanimonas cellulosilytica (strain DSM 15894 / CECT 5975 / LMG 20990 / XIL07) OX=446471 GN=Xcel_0480 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1156556 |
30 |
2724 |
336.1 |
35.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.42 ± 0.073 | 0.537 ± 0.01 |
6.441 ± 0.034 | 5.294 ± 0.036 |
2.654 ± 0.027 | 9.251 ± 0.031 |
2.126 ± 0.022 | 3.246 ± 0.032 |
1.613 ± 0.028 | 10.114 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.608 ± 0.019 | 1.639 ± 0.024 |
5.955 ± 0.038 | 2.719 ± 0.022 |
7.63 ± 0.052 | 4.843 ± 0.027 |
6.592 ± 0.04 | 9.926 ± 0.051 |
1.591 ± 0.019 | 1.8 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
