
Human associated porprismacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
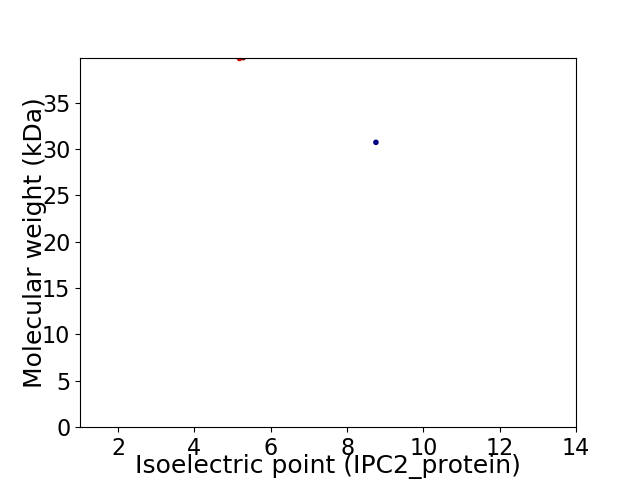
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
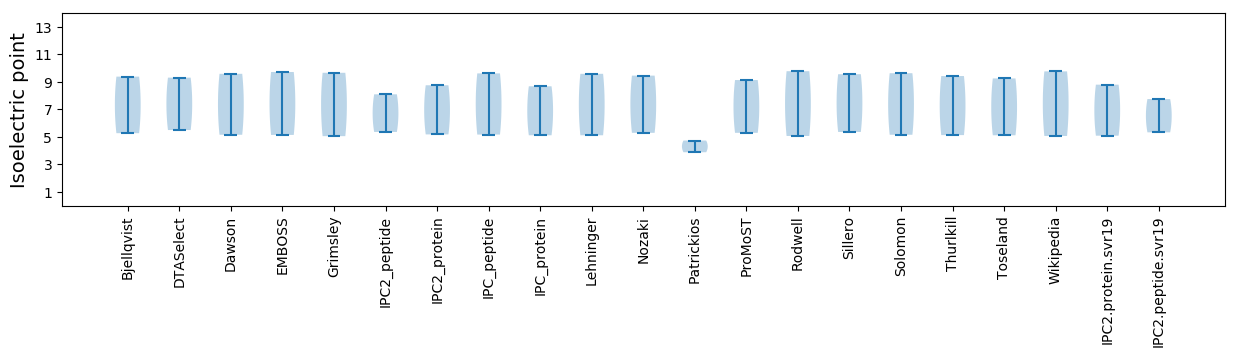
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5GSS8|A0A0B5GSS8_9VIRU Rep protein OS=Human associated porprismacovirus 2 OX=2170114 GN=rep PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 3.73VVILATNYY10 pKa = 10.61ASASYY15 pKa = 10.76QEE17 pKa = 4.29ILDD20 pKa = 3.88VNTVKK25 pKa = 11.08GNVTVIGIHH34 pKa = 5.93TPTGSTPVRR43 pKa = 11.84RR44 pKa = 11.84LSGFFDD50 pKa = 3.23QFRR53 pKa = 11.84KK54 pKa = 9.61FRR56 pKa = 11.84YY57 pKa = 8.93KK58 pKa = 9.37GCSVQVVPAATLPADD73 pKa = 3.8PLQVGFEE80 pKa = 4.13AGEE83 pKa = 3.95NTIDD87 pKa = 3.9MRR89 pKa = 11.84DD90 pKa = 3.58MLNPILFHH98 pKa = 5.87GTHH101 pKa = 6.44GEE103 pKa = 4.08SLQIAWNNIFVNPNRR118 pKa = 11.84YY119 pKa = 9.87DD120 pKa = 3.43GGADD124 pKa = 3.18TASYY128 pKa = 11.09SSGAGVDD135 pKa = 4.04ASDD138 pKa = 3.5LTFGNVTNSPVEE150 pKa = 3.94TQYY153 pKa = 11.47YY154 pKa = 8.63QAITDD159 pKa = 3.72RR160 pKa = 11.84SWRR163 pKa = 11.84KK164 pKa = 9.57FGIQSTFKK172 pKa = 10.76LPFMSPRR179 pKa = 11.84VWKK182 pKa = 10.6ASTLYY187 pKa = 10.14PIIPNSLQANGSMNTGNTTLPEE209 pKa = 4.29GFLPPGTYY217 pKa = 9.77FDD219 pKa = 4.72NYY221 pKa = 10.42DD222 pKa = 3.45GRR224 pKa = 11.84GLTMPIDD231 pKa = 4.53ADD233 pKa = 3.9QHH235 pKa = 6.91VLDD238 pKa = 4.51QKK240 pKa = 10.39WMSSGTTPLGWLPTVTMPRR259 pKa = 11.84VDD261 pKa = 4.08PNNGTSVVMAPRR273 pKa = 11.84TVLPKK278 pKa = 9.95IFMGILVLPPSYY290 pKa = 10.47LQEE293 pKa = 3.98LYY295 pKa = 10.34FRR297 pKa = 11.84VIITHH302 pKa = 5.5YY303 pKa = 10.91YY304 pKa = 7.78EE305 pKa = 4.68FKK307 pKa = 11.16GFTASLAPASVMPDD321 pKa = 2.78NKK323 pKa = 9.63GTYY326 pKa = 9.35EE327 pKa = 4.05NAVKK331 pKa = 8.75QTGTKK336 pKa = 9.46AASPEE341 pKa = 4.13EE342 pKa = 4.35VEE344 pKa = 4.5DD345 pKa = 3.94TTLEE349 pKa = 4.17SPTANVVRR357 pKa = 11.84ISDD360 pKa = 3.85GVFF363 pKa = 2.82
MM1 pKa = 7.86DD2 pKa = 3.73VVILATNYY10 pKa = 10.61ASASYY15 pKa = 10.76QEE17 pKa = 4.29ILDD20 pKa = 3.88VNTVKK25 pKa = 11.08GNVTVIGIHH34 pKa = 5.93TPTGSTPVRR43 pKa = 11.84RR44 pKa = 11.84LSGFFDD50 pKa = 3.23QFRR53 pKa = 11.84KK54 pKa = 9.61FRR56 pKa = 11.84YY57 pKa = 8.93KK58 pKa = 9.37GCSVQVVPAATLPADD73 pKa = 3.8PLQVGFEE80 pKa = 4.13AGEE83 pKa = 3.95NTIDD87 pKa = 3.9MRR89 pKa = 11.84DD90 pKa = 3.58MLNPILFHH98 pKa = 5.87GTHH101 pKa = 6.44GEE103 pKa = 4.08SLQIAWNNIFVNPNRR118 pKa = 11.84YY119 pKa = 9.87DD120 pKa = 3.43GGADD124 pKa = 3.18TASYY128 pKa = 11.09SSGAGVDD135 pKa = 4.04ASDD138 pKa = 3.5LTFGNVTNSPVEE150 pKa = 3.94TQYY153 pKa = 11.47YY154 pKa = 8.63QAITDD159 pKa = 3.72RR160 pKa = 11.84SWRR163 pKa = 11.84KK164 pKa = 9.57FGIQSTFKK172 pKa = 10.76LPFMSPRR179 pKa = 11.84VWKK182 pKa = 10.6ASTLYY187 pKa = 10.14PIIPNSLQANGSMNTGNTTLPEE209 pKa = 4.29GFLPPGTYY217 pKa = 9.77FDD219 pKa = 4.72NYY221 pKa = 10.42DD222 pKa = 3.45GRR224 pKa = 11.84GLTMPIDD231 pKa = 4.53ADD233 pKa = 3.9QHH235 pKa = 6.91VLDD238 pKa = 4.51QKK240 pKa = 10.39WMSSGTTPLGWLPTVTMPRR259 pKa = 11.84VDD261 pKa = 4.08PNNGTSVVMAPRR273 pKa = 11.84TVLPKK278 pKa = 9.95IFMGILVLPPSYY290 pKa = 10.47LQEE293 pKa = 3.98LYY295 pKa = 10.34FRR297 pKa = 11.84VIITHH302 pKa = 5.5YY303 pKa = 10.91YY304 pKa = 7.78EE305 pKa = 4.68FKK307 pKa = 11.16GFTASLAPASVMPDD321 pKa = 2.78NKK323 pKa = 9.63GTYY326 pKa = 9.35EE327 pKa = 4.05NAVKK331 pKa = 8.75QTGTKK336 pKa = 9.46AASPEE341 pKa = 4.13EE342 pKa = 4.35VEE344 pKa = 4.5DD345 pKa = 3.94TTLEE349 pKa = 4.17SPTANVVRR357 pKa = 11.84ISDD360 pKa = 3.85GVFF363 pKa = 2.82
Molecular weight: 39.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5GSS8|A0A0B5GSS8_9VIRU Rep protein OS=Human associated porprismacovirus 2 OX=2170114 GN=rep PE=4 SV=1
MM1 pKa = 6.35VQAWMITAPRR11 pKa = 11.84ANVSKK16 pKa = 10.49RR17 pKa = 11.84AIRR20 pKa = 11.84IMIDD24 pKa = 3.47KK25 pKa = 10.07NDD27 pKa = 3.64CKK29 pKa = 10.73KK30 pKa = 10.16WIIAKK35 pKa = 10.02EE36 pKa = 3.93KK37 pKa = 10.17GKK39 pKa = 10.3NGYY42 pKa = 7.38EE43 pKa = 3.45HH44 pKa = 5.56WQIRR48 pKa = 11.84IEE50 pKa = 4.12SSNTGFFEE58 pKa = 3.88WCKK61 pKa = 10.68LYY63 pKa = 10.51IPTAHH68 pKa = 6.08VEE70 pKa = 3.9KK71 pKa = 10.78AEE73 pKa = 4.15RR74 pKa = 11.84GVDD77 pKa = 3.37EE78 pKa = 4.63SRR80 pKa = 11.84YY81 pKa = 5.05EE82 pKa = 4.2TKK84 pKa = 9.96EE85 pKa = 3.77GKK87 pKa = 9.88YY88 pKa = 10.37VLYY91 pKa = 9.53TDD93 pKa = 3.42RR94 pKa = 11.84VEE96 pKa = 4.03ILKK99 pKa = 9.93QRR101 pKa = 11.84FGKK104 pKa = 8.87MRR106 pKa = 11.84PNQIRR111 pKa = 11.84ALEE114 pKa = 4.11ALEE117 pKa = 4.1ATNDD121 pKa = 3.59RR122 pKa = 11.84EE123 pKa = 4.21VLVWYY128 pKa = 10.07DD129 pKa = 3.09EE130 pKa = 4.29GGNVGKK136 pKa = 10.39SWLTGALWEE145 pKa = 4.84RR146 pKa = 11.84GLAYY150 pKa = 9.61YY151 pKa = 9.54VPPTVDD157 pKa = 2.8SVKK160 pKa = 11.22AMVQWVASCYY170 pKa = 9.73QSGGWRR176 pKa = 11.84PYY178 pKa = 10.81VIIDD182 pKa = 4.59IPRR185 pKa = 11.84SWKK188 pKa = 9.59WSEE191 pKa = 3.76QLYY194 pKa = 10.02VAIEE198 pKa = 4.26SIKK201 pKa = 11.03DD202 pKa = 3.15GLVYY206 pKa = 9.39DD207 pKa = 4.17TRR209 pKa = 11.84YY210 pKa = 9.55HH211 pKa = 5.6AQCINIRR218 pKa = 11.84GVKK221 pKa = 10.09VLVLTNTQPKK231 pKa = 9.91LDD233 pKa = 4.28KK234 pKa = 10.94LSADD238 pKa = 3.24RR239 pKa = 11.84WRR241 pKa = 11.84ICVFRR246 pKa = 11.84GRR248 pKa = 11.84RR249 pKa = 11.84AAGSALIVTLGADD262 pKa = 4.35FTPLL266 pKa = 3.22
MM1 pKa = 6.35VQAWMITAPRR11 pKa = 11.84ANVSKK16 pKa = 10.49RR17 pKa = 11.84AIRR20 pKa = 11.84IMIDD24 pKa = 3.47KK25 pKa = 10.07NDD27 pKa = 3.64CKK29 pKa = 10.73KK30 pKa = 10.16WIIAKK35 pKa = 10.02EE36 pKa = 3.93KK37 pKa = 10.17GKK39 pKa = 10.3NGYY42 pKa = 7.38EE43 pKa = 3.45HH44 pKa = 5.56WQIRR48 pKa = 11.84IEE50 pKa = 4.12SSNTGFFEE58 pKa = 3.88WCKK61 pKa = 10.68LYY63 pKa = 10.51IPTAHH68 pKa = 6.08VEE70 pKa = 3.9KK71 pKa = 10.78AEE73 pKa = 4.15RR74 pKa = 11.84GVDD77 pKa = 3.37EE78 pKa = 4.63SRR80 pKa = 11.84YY81 pKa = 5.05EE82 pKa = 4.2TKK84 pKa = 9.96EE85 pKa = 3.77GKK87 pKa = 9.88YY88 pKa = 10.37VLYY91 pKa = 9.53TDD93 pKa = 3.42RR94 pKa = 11.84VEE96 pKa = 4.03ILKK99 pKa = 9.93QRR101 pKa = 11.84FGKK104 pKa = 8.87MRR106 pKa = 11.84PNQIRR111 pKa = 11.84ALEE114 pKa = 4.11ALEE117 pKa = 4.1ATNDD121 pKa = 3.59RR122 pKa = 11.84EE123 pKa = 4.21VLVWYY128 pKa = 10.07DD129 pKa = 3.09EE130 pKa = 4.29GGNVGKK136 pKa = 10.39SWLTGALWEE145 pKa = 4.84RR146 pKa = 11.84GLAYY150 pKa = 9.61YY151 pKa = 9.54VPPTVDD157 pKa = 2.8SVKK160 pKa = 11.22AMVQWVASCYY170 pKa = 9.73QSGGWRR176 pKa = 11.84PYY178 pKa = 10.81VIIDD182 pKa = 4.59IPRR185 pKa = 11.84SWKK188 pKa = 9.59WSEE191 pKa = 3.76QLYY194 pKa = 10.02VAIEE198 pKa = 4.26SIKK201 pKa = 11.03DD202 pKa = 3.15GLVYY206 pKa = 9.39DD207 pKa = 4.17TRR209 pKa = 11.84YY210 pKa = 9.55HH211 pKa = 5.6AQCINIRR218 pKa = 11.84GVKK221 pKa = 10.09VLVLTNTQPKK231 pKa = 9.91LDD233 pKa = 4.28KK234 pKa = 10.94LSADD238 pKa = 3.24RR239 pKa = 11.84WRR241 pKa = 11.84ICVFRR246 pKa = 11.84GRR248 pKa = 11.84RR249 pKa = 11.84AAGSALIVTLGADD262 pKa = 4.35FTPLL266 pKa = 3.22
Molecular weight: 30.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
629 |
266 |
363 |
314.5 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.154 ± 0.481 | 0.954 ± 0.602 |
5.246 ± 0.233 | 4.769 ± 1.054 |
3.657 ± 1.155 | 7.472 ± 0.458 |
1.272 ± 0.094 | 6.041 ± 0.96 |
5.087 ± 1.58 | 6.836 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.432 | 4.769 ± 0.901 |
5.882 ± 1.624 | 3.498 ± 0.074 |
5.564 ± 1.515 | 6.2 ± 0.853 |
7.631 ± 1.783 | 8.267 ± 0.002 |
2.703 ± 1.175 | 4.452 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
