
Amaricoccus sp. HB172011
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Amaricoccus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
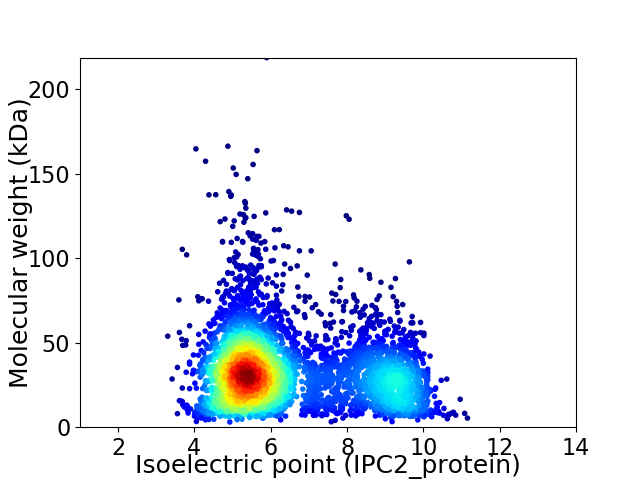
Virtual 2D-PAGE plot for 4522 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A501WPD3|A0A501WPD3_9RHOB Alpha/beta hydrolase OS=Amaricoccus sp. HB172011 OX=2589815 GN=FJM51_12510 PE=4 SV=1
MM1 pKa = 7.6IYY3 pKa = 8.85KK4 pKa = 8.34TARR7 pKa = 11.84GASLSTSGAPTSWIGAEE24 pKa = 3.4IGAYY28 pKa = 10.46LSGTSGNDD36 pKa = 2.94EE37 pKa = 4.79LYY39 pKa = 11.13GNDD42 pKa = 3.42SATLAGGLGDD52 pKa = 3.99DD53 pKa = 4.76TYY55 pKa = 10.88MVWVPSTHH63 pKa = 5.72VLEE66 pKa = 4.6AANAGIDD73 pKa = 4.37TLDD76 pKa = 3.08SWIWGTTVLPEE87 pKa = 3.9NFEE90 pKa = 4.44NLFLNGPGSTGGTGNASANIIAAGEE115 pKa = 4.11VGATLDD121 pKa = 3.85GLGGDD126 pKa = 4.37DD127 pKa = 4.14VLIGGAAADD136 pKa = 3.55IFRR139 pKa = 11.84IEE141 pKa = 4.52AGNGSDD147 pKa = 4.53AIVGFTPGSDD157 pKa = 3.37VIEE160 pKa = 4.98LIDD163 pKa = 3.55YY164 pKa = 8.39GFSNFAQVKK173 pKa = 10.17AIATKK178 pKa = 9.52TGKK181 pKa = 10.15DD182 pKa = 3.48LTLHH186 pKa = 6.92LAQDD190 pKa = 4.45EE191 pKa = 4.46ILVLRR196 pKa = 11.84DD197 pKa = 3.4TKK199 pKa = 10.74LAQLDD204 pKa = 4.55GYY206 pKa = 11.07DD207 pKa = 4.11FGFPPPAPAPGAGQGSLSGPGAVYY231 pKa = 9.88TDD233 pKa = 3.52HH234 pKa = 6.59GWYY237 pKa = 10.09VLNNAWNAGGLTYY250 pKa = 9.56GTDD253 pKa = 3.32YY254 pKa = 11.16RR255 pKa = 11.84LDD257 pKa = 4.51AIYY260 pKa = 10.82DD261 pKa = 4.0PDD263 pKa = 4.67DD264 pKa = 3.71LTTGVTFSWSFPRR277 pKa = 11.84TTQAYY282 pKa = 8.37PPILAYY288 pKa = 9.96PEE290 pKa = 5.0VIFGPAPMSGGHH302 pKa = 6.16KK303 pKa = 9.62VTDD306 pKa = 3.5TADD309 pKa = 3.87FFPLQVGEE317 pKa = 4.26IQDD320 pKa = 3.43LTAAYY325 pKa = 9.68DD326 pKa = 3.34VRR328 pKa = 11.84YY329 pKa = 10.48DD330 pKa = 4.97GNTDD334 pKa = 2.98WFNVAFDD341 pKa = 3.13IWLTDD346 pKa = 3.44VPNGGPSSVTNEE358 pKa = 3.53VMVWVHH364 pKa = 5.94QGAVKK369 pKa = 10.49PFGDD373 pKa = 3.86QVGTYY378 pKa = 10.43SDD380 pKa = 3.4GTIAAKK386 pKa = 10.01IHH388 pKa = 6.59SDD390 pKa = 4.24DD391 pKa = 4.29SSDD394 pKa = 2.96WTYY397 pKa = 10.23TAVVLDD403 pKa = 3.75SDD405 pKa = 4.56HH406 pKa = 6.93PVGTISITGILSEE419 pKa = 4.77LEE421 pKa = 3.93SLGIVSPDD429 pKa = 3.23EE430 pKa = 4.3YY431 pKa = 10.47IGSVEE436 pKa = 4.14LGSEE440 pKa = 4.05IVAGAGRR447 pKa = 11.84LTINEE452 pKa = 4.18LTLTARR458 pKa = 11.84LDD460 pKa = 3.6DD461 pKa = 3.89RR462 pKa = 11.84TLEE465 pKa = 4.08SSGAEE470 pKa = 4.0TTVTLLDD477 pKa = 3.82GAGTQTHH484 pKa = 6.48AADD487 pKa = 3.94TGWFF491 pKa = 3.58
MM1 pKa = 7.6IYY3 pKa = 8.85KK4 pKa = 8.34TARR7 pKa = 11.84GASLSTSGAPTSWIGAEE24 pKa = 3.4IGAYY28 pKa = 10.46LSGTSGNDD36 pKa = 2.94EE37 pKa = 4.79LYY39 pKa = 11.13GNDD42 pKa = 3.42SATLAGGLGDD52 pKa = 3.99DD53 pKa = 4.76TYY55 pKa = 10.88MVWVPSTHH63 pKa = 5.72VLEE66 pKa = 4.6AANAGIDD73 pKa = 4.37TLDD76 pKa = 3.08SWIWGTTVLPEE87 pKa = 3.9NFEE90 pKa = 4.44NLFLNGPGSTGGTGNASANIIAAGEE115 pKa = 4.11VGATLDD121 pKa = 3.85GLGGDD126 pKa = 4.37DD127 pKa = 4.14VLIGGAAADD136 pKa = 3.55IFRR139 pKa = 11.84IEE141 pKa = 4.52AGNGSDD147 pKa = 4.53AIVGFTPGSDD157 pKa = 3.37VIEE160 pKa = 4.98LIDD163 pKa = 3.55YY164 pKa = 8.39GFSNFAQVKK173 pKa = 10.17AIATKK178 pKa = 9.52TGKK181 pKa = 10.15DD182 pKa = 3.48LTLHH186 pKa = 6.92LAQDD190 pKa = 4.45EE191 pKa = 4.46ILVLRR196 pKa = 11.84DD197 pKa = 3.4TKK199 pKa = 10.74LAQLDD204 pKa = 4.55GYY206 pKa = 11.07DD207 pKa = 4.11FGFPPPAPAPGAGQGSLSGPGAVYY231 pKa = 9.88TDD233 pKa = 3.52HH234 pKa = 6.59GWYY237 pKa = 10.09VLNNAWNAGGLTYY250 pKa = 9.56GTDD253 pKa = 3.32YY254 pKa = 11.16RR255 pKa = 11.84LDD257 pKa = 4.51AIYY260 pKa = 10.82DD261 pKa = 4.0PDD263 pKa = 4.67DD264 pKa = 3.71LTTGVTFSWSFPRR277 pKa = 11.84TTQAYY282 pKa = 8.37PPILAYY288 pKa = 9.96PEE290 pKa = 5.0VIFGPAPMSGGHH302 pKa = 6.16KK303 pKa = 9.62VTDD306 pKa = 3.5TADD309 pKa = 3.87FFPLQVGEE317 pKa = 4.26IQDD320 pKa = 3.43LTAAYY325 pKa = 9.68DD326 pKa = 3.34VRR328 pKa = 11.84YY329 pKa = 10.48DD330 pKa = 4.97GNTDD334 pKa = 2.98WFNVAFDD341 pKa = 3.13IWLTDD346 pKa = 3.44VPNGGPSSVTNEE358 pKa = 3.53VMVWVHH364 pKa = 5.94QGAVKK369 pKa = 10.49PFGDD373 pKa = 3.86QVGTYY378 pKa = 10.43SDD380 pKa = 3.4GTIAAKK386 pKa = 10.01IHH388 pKa = 6.59SDD390 pKa = 4.24DD391 pKa = 4.29SSDD394 pKa = 2.96WTYY397 pKa = 10.23TAVVLDD403 pKa = 3.75SDD405 pKa = 4.56HH406 pKa = 6.93PVGTISITGILSEE419 pKa = 4.77LEE421 pKa = 3.93SLGIVSPDD429 pKa = 3.23EE430 pKa = 4.3YY431 pKa = 10.47IGSVEE436 pKa = 4.14LGSEE440 pKa = 4.05IVAGAGRR447 pKa = 11.84LTINEE452 pKa = 4.18LTLTARR458 pKa = 11.84LDD460 pKa = 3.6DD461 pKa = 3.89RR462 pKa = 11.84TLEE465 pKa = 4.08SSGAEE470 pKa = 4.0TTVTLLDD477 pKa = 3.82GAGTQTHH484 pKa = 6.48AADD487 pKa = 3.94TGWFF491 pKa = 3.58
Molecular weight: 51.28 kDa
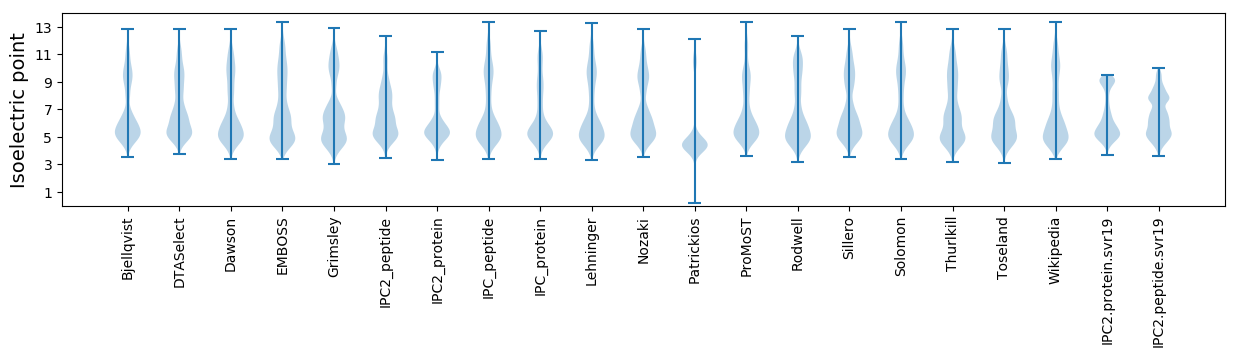
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A501WXA2|A0A501WXA2_9RHOB Uncharacterized protein OS=Amaricoccus sp. HB172011 OX=2589815 GN=FJM51_11985 PE=4 SV=1
MM1 pKa = 7.77ALTSSCVSRR10 pKa = 11.84ARR12 pKa = 11.84RR13 pKa = 11.84PARR16 pKa = 11.84PRR18 pKa = 11.84SSPAGRR24 pKa = 11.84RR25 pKa = 11.84GRR27 pKa = 11.84RR28 pKa = 11.84GARR31 pKa = 11.84PSGAARR37 pKa = 11.84SPSARR42 pKa = 11.84CRR44 pKa = 11.84ARR46 pKa = 11.84PGRR49 pKa = 11.84APSPRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PPPGAGPAPRR66 pKa = 11.84RR67 pKa = 11.84GPARR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84NN76 pKa = 3.01
MM1 pKa = 7.77ALTSSCVSRR10 pKa = 11.84ARR12 pKa = 11.84RR13 pKa = 11.84PARR16 pKa = 11.84PRR18 pKa = 11.84SSPAGRR24 pKa = 11.84RR25 pKa = 11.84GRR27 pKa = 11.84RR28 pKa = 11.84GARR31 pKa = 11.84PSGAARR37 pKa = 11.84SPSARR42 pKa = 11.84CRR44 pKa = 11.84ARR46 pKa = 11.84PGRR49 pKa = 11.84APSPRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PPPGAGPAPRR66 pKa = 11.84RR67 pKa = 11.84GPARR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84NN76 pKa = 3.01
Molecular weight: 8.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1433990 |
28 |
2028 |
317.1 |
34.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.117 ± 0.062 | 0.831 ± 0.012 |
5.59 ± 0.028 | 6.108 ± 0.037 |
3.549 ± 0.025 | 9.38 ± 0.044 |
1.912 ± 0.017 | 4.722 ± 0.025 |
2.361 ± 0.029 | 10.334 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.395 ± 0.017 | 2.13 ± 0.017 |
5.725 ± 0.027 | 2.347 ± 0.021 |
8.18 ± 0.039 | 4.625 ± 0.028 |
4.996 ± 0.025 | 7.175 ± 0.03 |
1.428 ± 0.014 | 2.095 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
