
Red clover powdery mildew-associated totivirus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; unclassified Totiviridae
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
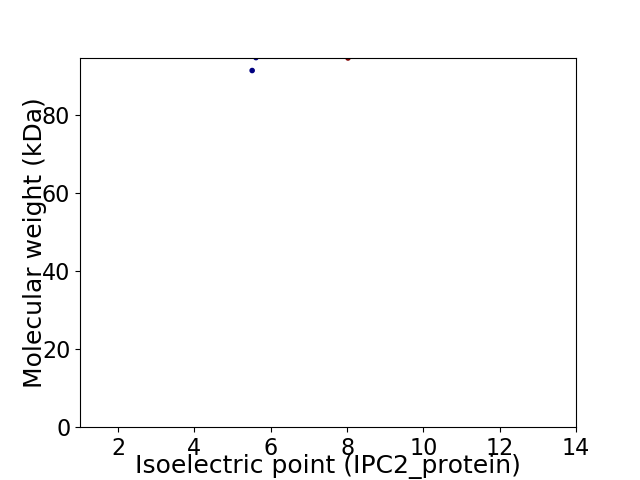
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
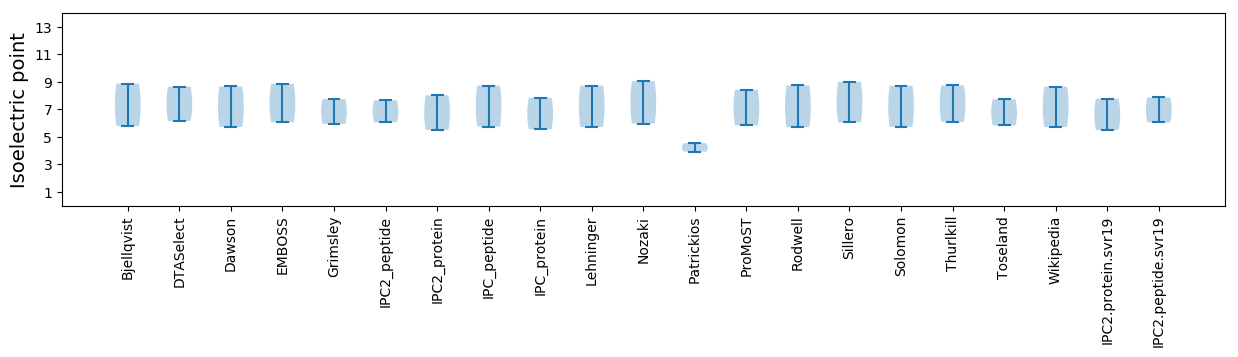
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
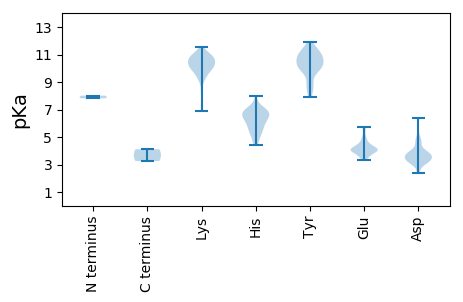
Protein with the lowest isoelectric point:
>tr|A0A0S3Q2D7|A0A0S3Q2D7_9VIRU RNA-directed RNA polymerase OS=Red clover powdery mildew-associated totivirus 5 OX=1714366 GN=RdRp PE=3 SV=1
MM1 pKa = 8.01EE2 pKa = 4.05YY3 pKa = 10.11TRR5 pKa = 11.84GLFNVDD11 pKa = 3.03GMIEE15 pKa = 4.2SFGDD19 pKa = 3.17GLIYY23 pKa = 10.61HH24 pKa = 6.35KK25 pKa = 9.75TRR27 pKa = 11.84LALNIADD34 pKa = 4.02YY35 pKa = 11.25NRR37 pKa = 11.84ADD39 pKa = 4.21FGTGDD44 pKa = 3.13IQMNRR49 pKa = 11.84HH50 pKa = 4.43STVNVRR56 pKa = 11.84SDD58 pKa = 3.34RR59 pKa = 11.84VSALAKK65 pKa = 10.1LSEE68 pKa = 4.51SNGEE72 pKa = 3.81KK73 pKa = 10.42FKK75 pKa = 10.91FRR77 pKa = 11.84RR78 pKa = 11.84LTNCLSQSTVYY89 pKa = 10.74GVVNTLRR96 pKa = 11.84STTPQHH102 pKa = 6.86SYY104 pKa = 11.25LGMSKK109 pKa = 10.43KK110 pKa = 10.63YY111 pKa = 8.9LTNEE115 pKa = 3.92GLPNEE120 pKa = 4.09QIIIKK125 pKa = 9.93RR126 pKa = 11.84LRR128 pKa = 11.84EE129 pKa = 3.97LGMHH133 pKa = 6.8HH134 pKa = 6.95DD135 pKa = 3.78VKK137 pKa = 10.66EE138 pKa = 3.87ARR140 pKa = 11.84FGQFYY145 pKa = 11.56NMLFTKK151 pKa = 10.08DD152 pKa = 3.75YY153 pKa = 8.61YY154 pKa = 11.55DD155 pKa = 3.75NMTALFAKK163 pKa = 10.22LHH165 pKa = 5.84LFSTRR170 pKa = 11.84LSIQEE175 pKa = 3.66RR176 pKa = 11.84MGVHH180 pKa = 6.35DD181 pKa = 4.85HH182 pKa = 7.26DD183 pKa = 4.75VVISTASLTKK193 pKa = 9.97MVGLANDD200 pKa = 4.49DD201 pKa = 3.3EE202 pKa = 4.51RR203 pKa = 11.84LAYY206 pKa = 9.9FKK208 pKa = 11.08SLLSSGTGRR217 pKa = 11.84LGFYY221 pKa = 9.56EE222 pKa = 3.75QDD224 pKa = 3.7YY225 pKa = 10.95NLEE228 pKa = 4.11TMLQNSIAWFNVNKK242 pKa = 9.99NDD244 pKa = 3.83PAARR248 pKa = 11.84EE249 pKa = 3.72YY250 pKa = 10.82RR251 pKa = 11.84QTVKK255 pKa = 9.74VWHH258 pKa = 6.41MYY260 pKa = 10.33KK261 pKa = 10.26YY262 pKa = 10.85DD263 pKa = 4.27DD264 pKa = 3.63GHH266 pKa = 5.97NVSGRR271 pKa = 11.84SFGSEE276 pKa = 3.67FGFDD280 pKa = 3.06GGKK283 pKa = 10.0FLIPEE288 pKa = 4.13CHH290 pKa = 5.22NTINYY295 pKa = 8.89IDD297 pKa = 3.93GVKK300 pKa = 10.0LVKK303 pKa = 10.73SRR305 pKa = 11.84DD306 pKa = 3.02NWFSDD311 pKa = 3.38EE312 pKa = 4.78NIDD315 pKa = 4.79KK316 pKa = 10.79FGSRR320 pKa = 11.84YY321 pKa = 10.12AGYY324 pKa = 10.77LNLSGMTEE332 pKa = 3.95LEE334 pKa = 3.86IAIVNHH340 pKa = 6.3ILLNTDD346 pKa = 2.39RR347 pKa = 11.84MTPFLCDD354 pKa = 3.26QEE356 pKa = 5.39LALCSDD362 pKa = 3.46SDD364 pKa = 3.78KK365 pKa = 11.21FLALASGEE373 pKa = 4.01ISTKK377 pKa = 7.64QTVVSSKK384 pKa = 7.27QVRR387 pKa = 11.84TVMSKK392 pKa = 10.59LAMNHH397 pKa = 6.16RR398 pKa = 11.84AQEE401 pKa = 4.11DD402 pKa = 3.81MLCAVRR408 pKa = 11.84SCKK411 pKa = 10.31YY412 pKa = 10.74LLAQPANEE420 pKa = 4.19TAEE423 pKa = 4.31SHH425 pKa = 4.84WWNVLPKK432 pKa = 10.16IVDD435 pKa = 3.99FPKK438 pKa = 10.2MGLKK442 pKa = 9.97RR443 pKa = 11.84AALHH447 pKa = 6.91FMLEE451 pKa = 4.23DD452 pKa = 3.3EE453 pKa = 4.96GVCISVDD460 pKa = 3.7SIKK463 pKa = 10.67YY464 pKa = 8.8ARR466 pKa = 11.84EE467 pKa = 3.79YY468 pKa = 10.99LSKK471 pKa = 10.5CDD473 pKa = 3.57RR474 pKa = 11.84AILEE478 pKa = 4.48SVFVNSCWFWAEE490 pKa = 3.57YY491 pKa = 8.73LTIYY495 pKa = 10.82NKK497 pKa = 9.87TNSYY501 pKa = 11.16DD502 pKa = 3.38LAAGLLRR509 pKa = 11.84GNDD512 pKa = 2.82SWLVEE517 pKa = 4.09RR518 pKa = 11.84EE519 pKa = 3.99RR520 pKa = 11.84PDD522 pKa = 3.03IMVSALIGRR531 pKa = 11.84PVPIAVHH538 pKa = 5.65SCCTTQWAKK547 pKa = 10.4PLSEE551 pKa = 4.04LARR554 pKa = 11.84LKK556 pKa = 10.92VCFGTISLTGRR567 pKa = 11.84DD568 pKa = 4.01DD569 pKa = 3.99YY570 pKa = 11.88NYY572 pKa = 9.78TLSDD576 pKa = 3.27KK577 pKa = 11.11DD578 pKa = 3.87VIFNSVVPPSGLALVTGLGGSLIQGTPYY606 pKa = 11.21GSMFGINQAVKK617 pKa = 9.5VRR619 pKa = 11.84KK620 pKa = 9.56GMRR623 pKa = 11.84TITSLNYY630 pKa = 9.87NDD632 pKa = 4.27LWALGVVSRR641 pKa = 11.84FNGYY645 pKa = 7.88DD646 pKa = 3.45LKK648 pKa = 11.1YY649 pKa = 10.23KK650 pKa = 10.53CPSSEE655 pKa = 4.11NVHH658 pKa = 6.54TIYY661 pKa = 10.82AANNVSIANPPVLFDD676 pKa = 4.44PEE678 pKa = 4.2ASNLSYY684 pKa = 10.72AIRR687 pKa = 11.84SITEE691 pKa = 3.39RR692 pKa = 11.84DD693 pKa = 3.2IVFGSDD699 pKa = 2.71TTTLLSSKK707 pKa = 10.68SVFYY711 pKa = 10.59WNRR714 pKa = 11.84FEE716 pKa = 4.4STTLEE721 pKa = 4.13SPKK724 pKa = 9.61WSAGIISNDD733 pKa = 3.03VEE735 pKa = 4.23EE736 pKa = 4.58AVLIRR741 pKa = 11.84GFEE744 pKa = 4.26CYY746 pKa = 10.36LDD748 pKa = 3.58EE749 pKa = 4.45TKK751 pKa = 10.54KK752 pKa = 9.27YY753 pKa = 8.19TVAISAEE760 pKa = 4.06YY761 pKa = 10.19DD762 pKa = 3.59VEE764 pKa = 4.47VSDD767 pKa = 4.05FHH769 pKa = 7.0VAKK772 pKa = 9.14LTAGIGLPTLDD783 pKa = 5.05GGLLLDD789 pKa = 4.58AEE791 pKa = 4.69NPAEE795 pKa = 4.14QTTEE799 pKa = 3.49LDD801 pKa = 3.62MPVLEE806 pKa = 5.46DD807 pKa = 3.4SALGAGSS814 pKa = 3.25
MM1 pKa = 8.01EE2 pKa = 4.05YY3 pKa = 10.11TRR5 pKa = 11.84GLFNVDD11 pKa = 3.03GMIEE15 pKa = 4.2SFGDD19 pKa = 3.17GLIYY23 pKa = 10.61HH24 pKa = 6.35KK25 pKa = 9.75TRR27 pKa = 11.84LALNIADD34 pKa = 4.02YY35 pKa = 11.25NRR37 pKa = 11.84ADD39 pKa = 4.21FGTGDD44 pKa = 3.13IQMNRR49 pKa = 11.84HH50 pKa = 4.43STVNVRR56 pKa = 11.84SDD58 pKa = 3.34RR59 pKa = 11.84VSALAKK65 pKa = 10.1LSEE68 pKa = 4.51SNGEE72 pKa = 3.81KK73 pKa = 10.42FKK75 pKa = 10.91FRR77 pKa = 11.84RR78 pKa = 11.84LTNCLSQSTVYY89 pKa = 10.74GVVNTLRR96 pKa = 11.84STTPQHH102 pKa = 6.86SYY104 pKa = 11.25LGMSKK109 pKa = 10.43KK110 pKa = 10.63YY111 pKa = 8.9LTNEE115 pKa = 3.92GLPNEE120 pKa = 4.09QIIIKK125 pKa = 9.93RR126 pKa = 11.84LRR128 pKa = 11.84EE129 pKa = 3.97LGMHH133 pKa = 6.8HH134 pKa = 6.95DD135 pKa = 3.78VKK137 pKa = 10.66EE138 pKa = 3.87ARR140 pKa = 11.84FGQFYY145 pKa = 11.56NMLFTKK151 pKa = 10.08DD152 pKa = 3.75YY153 pKa = 8.61YY154 pKa = 11.55DD155 pKa = 3.75NMTALFAKK163 pKa = 10.22LHH165 pKa = 5.84LFSTRR170 pKa = 11.84LSIQEE175 pKa = 3.66RR176 pKa = 11.84MGVHH180 pKa = 6.35DD181 pKa = 4.85HH182 pKa = 7.26DD183 pKa = 4.75VVISTASLTKK193 pKa = 9.97MVGLANDD200 pKa = 4.49DD201 pKa = 3.3EE202 pKa = 4.51RR203 pKa = 11.84LAYY206 pKa = 9.9FKK208 pKa = 11.08SLLSSGTGRR217 pKa = 11.84LGFYY221 pKa = 9.56EE222 pKa = 3.75QDD224 pKa = 3.7YY225 pKa = 10.95NLEE228 pKa = 4.11TMLQNSIAWFNVNKK242 pKa = 9.99NDD244 pKa = 3.83PAARR248 pKa = 11.84EE249 pKa = 3.72YY250 pKa = 10.82RR251 pKa = 11.84QTVKK255 pKa = 9.74VWHH258 pKa = 6.41MYY260 pKa = 10.33KK261 pKa = 10.26YY262 pKa = 10.85DD263 pKa = 4.27DD264 pKa = 3.63GHH266 pKa = 5.97NVSGRR271 pKa = 11.84SFGSEE276 pKa = 3.67FGFDD280 pKa = 3.06GGKK283 pKa = 10.0FLIPEE288 pKa = 4.13CHH290 pKa = 5.22NTINYY295 pKa = 8.89IDD297 pKa = 3.93GVKK300 pKa = 10.0LVKK303 pKa = 10.73SRR305 pKa = 11.84DD306 pKa = 3.02NWFSDD311 pKa = 3.38EE312 pKa = 4.78NIDD315 pKa = 4.79KK316 pKa = 10.79FGSRR320 pKa = 11.84YY321 pKa = 10.12AGYY324 pKa = 10.77LNLSGMTEE332 pKa = 3.95LEE334 pKa = 3.86IAIVNHH340 pKa = 6.3ILLNTDD346 pKa = 2.39RR347 pKa = 11.84MTPFLCDD354 pKa = 3.26QEE356 pKa = 5.39LALCSDD362 pKa = 3.46SDD364 pKa = 3.78KK365 pKa = 11.21FLALASGEE373 pKa = 4.01ISTKK377 pKa = 7.64QTVVSSKK384 pKa = 7.27QVRR387 pKa = 11.84TVMSKK392 pKa = 10.59LAMNHH397 pKa = 6.16RR398 pKa = 11.84AQEE401 pKa = 4.11DD402 pKa = 3.81MLCAVRR408 pKa = 11.84SCKK411 pKa = 10.31YY412 pKa = 10.74LLAQPANEE420 pKa = 4.19TAEE423 pKa = 4.31SHH425 pKa = 4.84WWNVLPKK432 pKa = 10.16IVDD435 pKa = 3.99FPKK438 pKa = 10.2MGLKK442 pKa = 9.97RR443 pKa = 11.84AALHH447 pKa = 6.91FMLEE451 pKa = 4.23DD452 pKa = 3.3EE453 pKa = 4.96GVCISVDD460 pKa = 3.7SIKK463 pKa = 10.67YY464 pKa = 8.8ARR466 pKa = 11.84EE467 pKa = 3.79YY468 pKa = 10.99LSKK471 pKa = 10.5CDD473 pKa = 3.57RR474 pKa = 11.84AILEE478 pKa = 4.48SVFVNSCWFWAEE490 pKa = 3.57YY491 pKa = 8.73LTIYY495 pKa = 10.82NKK497 pKa = 9.87TNSYY501 pKa = 11.16DD502 pKa = 3.38LAAGLLRR509 pKa = 11.84GNDD512 pKa = 2.82SWLVEE517 pKa = 4.09RR518 pKa = 11.84EE519 pKa = 3.99RR520 pKa = 11.84PDD522 pKa = 3.03IMVSALIGRR531 pKa = 11.84PVPIAVHH538 pKa = 5.65SCCTTQWAKK547 pKa = 10.4PLSEE551 pKa = 4.04LARR554 pKa = 11.84LKK556 pKa = 10.92VCFGTISLTGRR567 pKa = 11.84DD568 pKa = 4.01DD569 pKa = 3.99YY570 pKa = 11.88NYY572 pKa = 9.78TLSDD576 pKa = 3.27KK577 pKa = 11.11DD578 pKa = 3.87VIFNSVVPPSGLALVTGLGGSLIQGTPYY606 pKa = 11.21GSMFGINQAVKK617 pKa = 9.5VRR619 pKa = 11.84KK620 pKa = 9.56GMRR623 pKa = 11.84TITSLNYY630 pKa = 9.87NDD632 pKa = 4.27LWALGVVSRR641 pKa = 11.84FNGYY645 pKa = 7.88DD646 pKa = 3.45LKK648 pKa = 11.1YY649 pKa = 10.23KK650 pKa = 10.53CPSSEE655 pKa = 4.11NVHH658 pKa = 6.54TIYY661 pKa = 10.82AANNVSIANPPVLFDD676 pKa = 4.44PEE678 pKa = 4.2ASNLSYY684 pKa = 10.72AIRR687 pKa = 11.84SITEE691 pKa = 3.39RR692 pKa = 11.84DD693 pKa = 3.2IVFGSDD699 pKa = 2.71TTTLLSSKK707 pKa = 10.68SVFYY711 pKa = 10.59WNRR714 pKa = 11.84FEE716 pKa = 4.4STTLEE721 pKa = 4.13SPKK724 pKa = 9.61WSAGIISNDD733 pKa = 3.03VEE735 pKa = 4.23EE736 pKa = 4.58AVLIRR741 pKa = 11.84GFEE744 pKa = 4.26CYY746 pKa = 10.36LDD748 pKa = 3.58EE749 pKa = 4.45TKK751 pKa = 10.54KK752 pKa = 9.27YY753 pKa = 8.19TVAISAEE760 pKa = 4.06YY761 pKa = 10.19DD762 pKa = 3.59VEE764 pKa = 4.47VSDD767 pKa = 4.05FHH769 pKa = 7.0VAKK772 pKa = 9.14LTAGIGLPTLDD783 pKa = 5.05GGLLLDD789 pKa = 4.58AEE791 pKa = 4.69NPAEE795 pKa = 4.14QTTEE799 pKa = 3.49LDD801 pKa = 3.62MPVLEE806 pKa = 5.46DD807 pKa = 3.4SALGAGSS814 pKa = 3.25
Molecular weight: 91.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3Q2D7|A0A0S3Q2D7_9VIRU RNA-directed RNA polymerase OS=Red clover powdery mildew-associated totivirus 5 OX=1714366 GN=RdRp PE=3 SV=1
MM1 pKa = 7.81PYY3 pKa = 10.36VPTFLAPTDD12 pKa = 3.45SSRR15 pKa = 11.84VFRR18 pKa = 11.84EE19 pKa = 3.36VDD21 pKa = 3.86FNEE24 pKa = 3.99ADD26 pKa = 3.56LVLFEE31 pKa = 4.96CLSGLEE37 pKa = 4.71LDD39 pKa = 4.77GSMHH43 pKa = 6.96LHH45 pKa = 5.45IQEE48 pKa = 4.13SAVPIIAVYY57 pKa = 9.82LPKK60 pKa = 10.83VKK62 pKa = 9.28VTACYY67 pKa = 10.25ISIDD71 pKa = 3.77YY72 pKa = 9.9QIKK75 pKa = 8.36HH76 pKa = 5.27TNKK79 pKa = 10.16DD80 pKa = 3.08VLLRR84 pKa = 11.84FSRR87 pKa = 11.84MQYY90 pKa = 10.96GPDD93 pKa = 3.66LFPFGLVSDD102 pKa = 4.65LDD104 pKa = 3.46IVRR107 pKa = 11.84YY108 pKa = 10.13AFFISRR114 pKa = 11.84KK115 pKa = 6.91TIKK118 pKa = 9.83KK119 pKa = 10.01RR120 pKa = 11.84KK121 pKa = 8.96DD122 pKa = 3.3YY123 pKa = 10.45KK124 pKa = 10.86IKK126 pKa = 10.49DD127 pKa = 3.62FPIIDD132 pKa = 3.24SWFRR136 pKa = 11.84GSTEE140 pKa = 3.87PPVSKK145 pKa = 10.84VSVNHH150 pKa = 5.79LRR152 pKa = 11.84HH153 pKa = 4.7MTMAEE158 pKa = 3.78VRR160 pKa = 11.84LMGRR164 pKa = 11.84GVIEE168 pKa = 4.86DD169 pKa = 3.54KK170 pKa = 11.07VSYY173 pKa = 10.29ILPFLSRR180 pKa = 11.84MAKK183 pKa = 10.0VGMSEE188 pKa = 3.94AMFIGIVLWANSLPTQQHH206 pKa = 5.58EE207 pKa = 4.29LCKK210 pKa = 10.82LSGLWDD216 pKa = 3.9LSFTSMEE223 pKa = 4.16DD224 pKa = 3.03FATKK228 pKa = 10.24VKK230 pKa = 10.81ARR232 pKa = 11.84FSLRR236 pKa = 11.84LKK238 pKa = 10.48ALQNLVSVDD247 pKa = 3.22LHH249 pKa = 6.71CFFEE253 pKa = 5.57LEE255 pKa = 4.2VLVNRR260 pKa = 11.84GVGEE264 pKa = 4.23LNWEE268 pKa = 4.03KK269 pKa = 11.16EE270 pKa = 3.95KK271 pKa = 10.86MNRR274 pKa = 11.84TRR276 pKa = 11.84PNLAQFKK283 pKa = 9.32PEE285 pKa = 3.89QVFSKK290 pKa = 10.53ACSLFTDD297 pKa = 3.77ILRR300 pKa = 11.84RR301 pKa = 11.84GGRR304 pKa = 11.84PKK306 pKa = 10.2KK307 pKa = 10.52RR308 pKa = 11.84NWSEE312 pKa = 3.53FWSNRR317 pKa = 11.84WEE319 pKa = 3.98WAPTGSSHH327 pKa = 6.18SQYY330 pKa = 11.54AEE332 pKa = 3.9DD333 pKa = 5.31DD334 pKa = 4.1VYY336 pKa = 11.44KK337 pKa = 11.07SKK339 pKa = 11.49DD340 pKa = 3.05PFNRR344 pKa = 11.84HH345 pKa = 5.63KK346 pKa = 11.06LFTLCSMPEE355 pKa = 3.47RR356 pKa = 11.84SFEE359 pKa = 4.1YY360 pKa = 9.93FLKK363 pKa = 10.63RR364 pKa = 11.84KK365 pKa = 9.47PEE367 pKa = 4.33TIAWTSTKK375 pKa = 10.56YY376 pKa = 10.29EE377 pKa = 4.04WTKK380 pKa = 9.96MRR382 pKa = 11.84AIYY385 pKa = 10.14GVDD388 pKa = 2.52ITNFIMTTFAMGDD401 pKa = 3.84CEE403 pKa = 5.59DD404 pKa = 3.88VLSSKK409 pKa = 10.51FPIGKK414 pKa = 9.2AANEE418 pKa = 4.11VNVRR422 pKa = 11.84STVKK426 pKa = 9.39TLLGNGVPFCFDD438 pKa = 3.37FEE440 pKa = 5.02DD441 pKa = 4.71FNSQHH446 pKa = 6.7SIEE449 pKa = 5.01SMVQVIRR456 pKa = 11.84AYY458 pKa = 10.32SSTFKK463 pKa = 10.07MHH465 pKa = 6.86MSEE468 pKa = 3.98QQYY471 pKa = 10.66NAIDD475 pKa = 3.5WVCRR479 pKa = 11.84SFYY482 pKa = 11.04DD483 pKa = 3.83SFVVEE488 pKa = 5.0DD489 pKa = 3.57NRR491 pKa = 11.84GDD493 pKa = 3.79KK494 pKa = 11.01DD495 pKa = 3.47KK496 pKa = 11.52GPCNSSASSRR506 pKa = 11.84EE507 pKa = 3.5RR508 pKa = 11.84AKK510 pKa = 10.11MYY512 pKa = 9.74RR513 pKa = 11.84TMGTLLSGCRR523 pKa = 11.84LTTFLNTVLNYY534 pKa = 9.92IYY536 pKa = 11.09VNLSIEE542 pKa = 4.26GDD544 pKa = 3.82DD545 pKa = 6.35LIATHH550 pKa = 6.59NGDD553 pKa = 4.92DD554 pKa = 3.38ILAVVSSYY562 pKa = 11.45RR563 pKa = 11.84EE564 pKa = 3.63VQAISRR570 pKa = 11.84GAEE573 pKa = 3.38RR574 pKa = 11.84HH575 pKa = 5.2QIRR578 pKa = 11.84FQPSKK583 pKa = 10.88CFLGSVAEE591 pKa = 4.17FLRR594 pKa = 11.84IDD596 pKa = 3.48HH597 pKa = 6.79HH598 pKa = 7.95SGAGGQYY605 pKa = 8.12LTRR608 pKa = 11.84AISTLVHH615 pKa = 6.77GPTEE619 pKa = 3.89MAVPNNLVAQLTAINTRR636 pKa = 11.84ANEE639 pKa = 3.89AKK641 pKa = 10.64ARR643 pKa = 11.84GADD646 pKa = 3.23HH647 pKa = 7.03AFIAGVVKK655 pKa = 10.67SQLPYY660 pKa = 10.42LCKK663 pKa = 9.99VWQITEE669 pKa = 4.32DD670 pKa = 3.42EE671 pKa = 4.49VEE673 pKa = 4.48AVFSTHH679 pKa = 6.21LCFGGLSNEE688 pKa = 4.04ITPKK692 pKa = 10.49SLSKK696 pKa = 10.83SIILEE701 pKa = 3.83EE702 pKa = 5.74DD703 pKa = 2.42ITANKK708 pKa = 9.37RR709 pKa = 11.84PSVLADD715 pKa = 3.21EE716 pKa = 5.73GRR718 pKa = 11.84FFPGAFSYY726 pKa = 10.87AKK728 pKa = 10.04SLTSTIIPEE737 pKa = 4.21SYY739 pKa = 9.77FGKK742 pKa = 10.29VEE744 pKa = 4.04HH745 pKa = 6.39AVSRR749 pKa = 11.84TVTALSCSRR758 pKa = 11.84RR759 pKa = 11.84FRR761 pKa = 11.84LVVKK765 pKa = 10.24DD766 pKa = 3.62NKK768 pKa = 10.14TPGVTDD774 pKa = 3.59YY775 pKa = 11.23IKK777 pKa = 10.9ANRR780 pKa = 11.84YY781 pKa = 9.17GLFRR785 pKa = 11.84SKK787 pKa = 9.69LTAGKK792 pKa = 8.27ITLARR797 pKa = 11.84AFNIPIIDD805 pKa = 3.3IKK807 pKa = 9.71YY808 pKa = 8.46QKK810 pKa = 10.34QEE812 pKa = 3.69ILEE815 pKa = 4.21ILASEE820 pKa = 4.05QDD822 pKa = 3.12KK823 pKa = 10.93MEE825 pKa = 4.56ALRR828 pKa = 11.84ILFF831 pKa = 4.11
MM1 pKa = 7.81PYY3 pKa = 10.36VPTFLAPTDD12 pKa = 3.45SSRR15 pKa = 11.84VFRR18 pKa = 11.84EE19 pKa = 3.36VDD21 pKa = 3.86FNEE24 pKa = 3.99ADD26 pKa = 3.56LVLFEE31 pKa = 4.96CLSGLEE37 pKa = 4.71LDD39 pKa = 4.77GSMHH43 pKa = 6.96LHH45 pKa = 5.45IQEE48 pKa = 4.13SAVPIIAVYY57 pKa = 9.82LPKK60 pKa = 10.83VKK62 pKa = 9.28VTACYY67 pKa = 10.25ISIDD71 pKa = 3.77YY72 pKa = 9.9QIKK75 pKa = 8.36HH76 pKa = 5.27TNKK79 pKa = 10.16DD80 pKa = 3.08VLLRR84 pKa = 11.84FSRR87 pKa = 11.84MQYY90 pKa = 10.96GPDD93 pKa = 3.66LFPFGLVSDD102 pKa = 4.65LDD104 pKa = 3.46IVRR107 pKa = 11.84YY108 pKa = 10.13AFFISRR114 pKa = 11.84KK115 pKa = 6.91TIKK118 pKa = 9.83KK119 pKa = 10.01RR120 pKa = 11.84KK121 pKa = 8.96DD122 pKa = 3.3YY123 pKa = 10.45KK124 pKa = 10.86IKK126 pKa = 10.49DD127 pKa = 3.62FPIIDD132 pKa = 3.24SWFRR136 pKa = 11.84GSTEE140 pKa = 3.87PPVSKK145 pKa = 10.84VSVNHH150 pKa = 5.79LRR152 pKa = 11.84HH153 pKa = 4.7MTMAEE158 pKa = 3.78VRR160 pKa = 11.84LMGRR164 pKa = 11.84GVIEE168 pKa = 4.86DD169 pKa = 3.54KK170 pKa = 11.07VSYY173 pKa = 10.29ILPFLSRR180 pKa = 11.84MAKK183 pKa = 10.0VGMSEE188 pKa = 3.94AMFIGIVLWANSLPTQQHH206 pKa = 5.58EE207 pKa = 4.29LCKK210 pKa = 10.82LSGLWDD216 pKa = 3.9LSFTSMEE223 pKa = 4.16DD224 pKa = 3.03FATKK228 pKa = 10.24VKK230 pKa = 10.81ARR232 pKa = 11.84FSLRR236 pKa = 11.84LKK238 pKa = 10.48ALQNLVSVDD247 pKa = 3.22LHH249 pKa = 6.71CFFEE253 pKa = 5.57LEE255 pKa = 4.2VLVNRR260 pKa = 11.84GVGEE264 pKa = 4.23LNWEE268 pKa = 4.03KK269 pKa = 11.16EE270 pKa = 3.95KK271 pKa = 10.86MNRR274 pKa = 11.84TRR276 pKa = 11.84PNLAQFKK283 pKa = 9.32PEE285 pKa = 3.89QVFSKK290 pKa = 10.53ACSLFTDD297 pKa = 3.77ILRR300 pKa = 11.84RR301 pKa = 11.84GGRR304 pKa = 11.84PKK306 pKa = 10.2KK307 pKa = 10.52RR308 pKa = 11.84NWSEE312 pKa = 3.53FWSNRR317 pKa = 11.84WEE319 pKa = 3.98WAPTGSSHH327 pKa = 6.18SQYY330 pKa = 11.54AEE332 pKa = 3.9DD333 pKa = 5.31DD334 pKa = 4.1VYY336 pKa = 11.44KK337 pKa = 11.07SKK339 pKa = 11.49DD340 pKa = 3.05PFNRR344 pKa = 11.84HH345 pKa = 5.63KK346 pKa = 11.06LFTLCSMPEE355 pKa = 3.47RR356 pKa = 11.84SFEE359 pKa = 4.1YY360 pKa = 9.93FLKK363 pKa = 10.63RR364 pKa = 11.84KK365 pKa = 9.47PEE367 pKa = 4.33TIAWTSTKK375 pKa = 10.56YY376 pKa = 10.29EE377 pKa = 4.04WTKK380 pKa = 9.96MRR382 pKa = 11.84AIYY385 pKa = 10.14GVDD388 pKa = 2.52ITNFIMTTFAMGDD401 pKa = 3.84CEE403 pKa = 5.59DD404 pKa = 3.88VLSSKK409 pKa = 10.51FPIGKK414 pKa = 9.2AANEE418 pKa = 4.11VNVRR422 pKa = 11.84STVKK426 pKa = 9.39TLLGNGVPFCFDD438 pKa = 3.37FEE440 pKa = 5.02DD441 pKa = 4.71FNSQHH446 pKa = 6.7SIEE449 pKa = 5.01SMVQVIRR456 pKa = 11.84AYY458 pKa = 10.32SSTFKK463 pKa = 10.07MHH465 pKa = 6.86MSEE468 pKa = 3.98QQYY471 pKa = 10.66NAIDD475 pKa = 3.5WVCRR479 pKa = 11.84SFYY482 pKa = 11.04DD483 pKa = 3.83SFVVEE488 pKa = 5.0DD489 pKa = 3.57NRR491 pKa = 11.84GDD493 pKa = 3.79KK494 pKa = 11.01DD495 pKa = 3.47KK496 pKa = 11.52GPCNSSASSRR506 pKa = 11.84EE507 pKa = 3.5RR508 pKa = 11.84AKK510 pKa = 10.11MYY512 pKa = 9.74RR513 pKa = 11.84TMGTLLSGCRR523 pKa = 11.84LTTFLNTVLNYY534 pKa = 9.92IYY536 pKa = 11.09VNLSIEE542 pKa = 4.26GDD544 pKa = 3.82DD545 pKa = 6.35LIATHH550 pKa = 6.59NGDD553 pKa = 4.92DD554 pKa = 3.38ILAVVSSYY562 pKa = 11.45RR563 pKa = 11.84EE564 pKa = 3.63VQAISRR570 pKa = 11.84GAEE573 pKa = 3.38RR574 pKa = 11.84HH575 pKa = 5.2QIRR578 pKa = 11.84FQPSKK583 pKa = 10.88CFLGSVAEE591 pKa = 4.17FLRR594 pKa = 11.84IDD596 pKa = 3.48HH597 pKa = 6.79HH598 pKa = 7.95SGAGGQYY605 pKa = 8.12LTRR608 pKa = 11.84AISTLVHH615 pKa = 6.77GPTEE619 pKa = 3.89MAVPNNLVAQLTAINTRR636 pKa = 11.84ANEE639 pKa = 3.89AKK641 pKa = 10.64ARR643 pKa = 11.84GADD646 pKa = 3.23HH647 pKa = 7.03AFIAGVVKK655 pKa = 10.67SQLPYY660 pKa = 10.42LCKK663 pKa = 9.99VWQITEE669 pKa = 4.32DD670 pKa = 3.42EE671 pKa = 4.49VEE673 pKa = 4.48AVFSTHH679 pKa = 6.21LCFGGLSNEE688 pKa = 4.04ITPKK692 pKa = 10.49SLSKK696 pKa = 10.83SIILEE701 pKa = 3.83EE702 pKa = 5.74DD703 pKa = 2.42ITANKK708 pKa = 9.37RR709 pKa = 11.84PSVLADD715 pKa = 3.21EE716 pKa = 5.73GRR718 pKa = 11.84FFPGAFSYY726 pKa = 10.87AKK728 pKa = 10.04SLTSTIIPEE737 pKa = 4.21SYY739 pKa = 9.77FGKK742 pKa = 10.29VEE744 pKa = 4.04HH745 pKa = 6.39AVSRR749 pKa = 11.84TVTALSCSRR758 pKa = 11.84RR759 pKa = 11.84FRR761 pKa = 11.84LVVKK765 pKa = 10.24DD766 pKa = 3.62NKK768 pKa = 10.14TPGVTDD774 pKa = 3.59YY775 pKa = 11.23IKK777 pKa = 10.9ANRR780 pKa = 11.84YY781 pKa = 9.17GLFRR785 pKa = 11.84SKK787 pKa = 9.69LTAGKK792 pKa = 8.27ITLARR797 pKa = 11.84AFNIPIIDD805 pKa = 3.3IKK807 pKa = 9.71YY808 pKa = 8.46QKK810 pKa = 10.34QEE812 pKa = 3.69ILEE815 pKa = 4.21ILASEE820 pKa = 4.05QDD822 pKa = 3.12KK823 pKa = 10.93MEE825 pKa = 4.56ALRR828 pKa = 11.84ILFF831 pKa = 4.11
Molecular weight: 94.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1645 |
814 |
831 |
822.5 |
92.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.626 ± 0.006 | 1.763 ± 0.034 |
5.714 ± 0.439 | 5.836 ± 0.147 |
5.228 ± 0.739 | 5.836 ± 0.635 |
2.249 ± 0.03 | 5.593 ± 0.443 |
5.836 ± 0.538 | 9.422 ± 0.714 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.675 ± 0.022 | 4.863 ± 0.725 |
3.343 ± 0.412 | 2.492 ± 0.224 |
5.653 ± 0.491 | 8.754 ± 0.025 |
5.897 ± 0.196 | 6.93 ± 0.138 |
1.459 ± 0.012 | 3.83 ± 0.472 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
