
Methylocella silvestris (strain DSM 15510 / CIP 108128 / LMG 27833 / NCIMB 13906 / BL2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Beijerinckiaceae; Methylocella; Methylocella silvestris
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
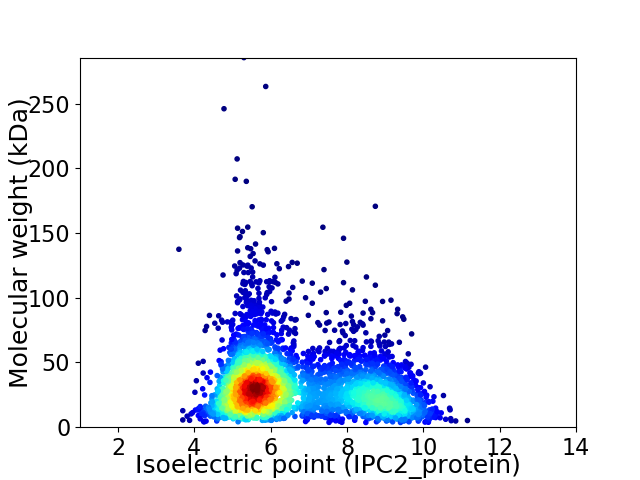
Virtual 2D-PAGE plot for 3816 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
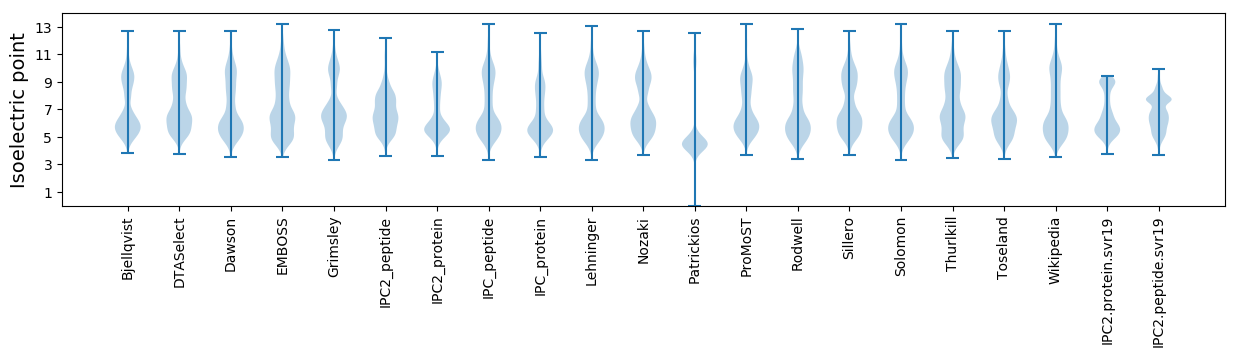
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8EQ36|B8EQ36_METSB ABC transporter related OS=Methylocella silvestris (strain DSM 15510 / CIP 108128 / LMG 27833 / NCIMB 13906 / BL2) OX=395965 GN=Msil_2603 PE=3 SV=1
MM1 pKa = 7.64SGITFIVEE9 pKa = 4.07GATGLDD15 pKa = 3.46EE16 pKa = 4.89QDD18 pKa = 3.39VKK20 pKa = 11.58NSFEE24 pKa = 4.12NLRR27 pKa = 11.84NASGDD32 pKa = 3.37FDD34 pKa = 5.06QDD36 pKa = 2.84ILYY39 pKa = 9.77IASKK43 pKa = 9.3TRR45 pKa = 11.84VLIIIANDD53 pKa = 3.8TVSGTSATYY62 pKa = 9.73PVTSGDD68 pKa = 3.03TAYY71 pKa = 10.96YY72 pKa = 8.45GTYY75 pKa = 9.89DD76 pKa = 3.53YY77 pKa = 11.08VIRR80 pKa = 11.84IPKK83 pKa = 9.84QGFDD87 pKa = 3.3GDD89 pKa = 4.4TYY91 pKa = 11.64NHH93 pKa = 6.52VGTDD97 pKa = 2.81KK98 pKa = 10.76WSLEE102 pKa = 3.53RR103 pKa = 11.84LYY105 pKa = 10.93AHH107 pKa = 6.94EE108 pKa = 4.46VAHH111 pKa = 5.9VMQAIGDD118 pKa = 4.12NAPSTQAEE126 pKa = 4.28RR127 pKa = 11.84EE128 pKa = 4.13EE129 pKa = 4.2QATKK133 pKa = 10.67YY134 pKa = 10.17SNHH137 pKa = 6.11IMSTAHH143 pKa = 6.88PEE145 pKa = 4.03EE146 pKa = 4.88SPQTHH151 pKa = 6.47YY152 pKa = 11.64YY153 pKa = 8.02NTEE156 pKa = 3.76GDD158 pKa = 3.58TGSGLNQNSGSGDD171 pKa = 3.62SPHH174 pKa = 7.97ILDD177 pKa = 4.81DD178 pKa = 4.3SSPLSEE184 pKa = 4.67NLPGAPLPSQLAAIQPAFGDD204 pKa = 4.7AINASSPLVIDD215 pKa = 4.66LSSGHH220 pKa = 6.31SGVTLTTWSASTTEE234 pKa = 4.41TYY236 pKa = 10.87FDD238 pKa = 4.88LDD240 pKa = 4.05DD241 pKa = 4.68NGFAVQTAWVSGDD254 pKa = 3.18TGLLARR260 pKa = 11.84DD261 pKa = 3.68LNEE264 pKa = 3.99NGIIDD269 pKa = 3.83SSAEE273 pKa = 3.95LFGSPTVDD281 pKa = 3.15GFAKK285 pKa = 10.62LAMLDD290 pKa = 3.84SNHH293 pKa = 7.22DD294 pKa = 3.74LRR296 pKa = 11.84IDD298 pKa = 3.96SNDD301 pKa = 3.78DD302 pKa = 2.98AWDD305 pKa = 3.8SLVVWNDD312 pKa = 3.23DD313 pKa = 3.24GDD315 pKa = 4.47AVTQSGEE322 pKa = 4.14LHH324 pKa = 6.37SLASLNIANIDD335 pKa = 3.77LAGVASSTSTISGNSISHH353 pKa = 5.23VSKK356 pKa = 9.69ITFTSGATAAIDD368 pKa = 3.8DD369 pKa = 3.91AWFVHH374 pKa = 7.23DD375 pKa = 4.67NTNSFYY381 pKa = 10.45TGEE384 pKa = 4.13YY385 pKa = 9.05TLDD388 pKa = 3.34VDD390 pKa = 4.06TLFLPRR396 pKa = 11.84VRR398 pKa = 11.84GYY400 pKa = 8.97GTLPDD405 pKa = 3.66LTIAMSLDD413 pKa = 3.57SDD415 pKa = 4.81LKK417 pKa = 11.3DD418 pKa = 3.57LVLGFAGTDD427 pKa = 3.22LSEE430 pKa = 4.23FADD433 pKa = 3.83AKK435 pKa = 10.07TAITDD440 pKa = 4.07ILYY443 pKa = 9.87KK444 pKa = 8.91WAGVEE449 pKa = 4.67GVDD452 pKa = 4.12PDD454 pKa = 4.63SRR456 pKa = 11.84GIWVDD461 pKa = 3.34AQHH464 pKa = 7.13LAFLEE469 pKa = 4.06QFIGSDD475 pKa = 3.48FLQITTHH482 pKa = 6.51SADD485 pKa = 3.58PQPWAGALIEE495 pKa = 4.81DD496 pKa = 5.12AYY498 pKa = 10.94QAAFAMLSADD508 pKa = 3.36ILVQTGFGQLFDD520 pKa = 3.55TAITYY525 pKa = 9.75NPSSGTRR532 pKa = 11.84TGDD535 pKa = 3.14TDD537 pKa = 3.93LSEE540 pKa = 5.05DD541 pKa = 4.59AIDD544 pKa = 4.56DD545 pKa = 4.15LVSIAPSSGPEE556 pKa = 3.71NEE558 pKa = 4.44AFWVSVAWALDD569 pKa = 3.52VTKK572 pKa = 10.81GISNLSYY579 pKa = 11.42DD580 pKa = 3.69EE581 pKa = 4.38TTWLSDD587 pKa = 4.79AITASNALSDD597 pKa = 3.8WATVLHH603 pKa = 6.74SYY605 pKa = 10.64DD606 pKa = 3.81PSAGTASQNGTNGAEE621 pKa = 4.06TLYY624 pKa = 11.42GDD626 pKa = 3.57VTADD630 pKa = 3.87TIHH633 pKa = 6.6GFNGADD639 pKa = 3.82TIYY642 pKa = 11.06GDD644 pKa = 3.8YY645 pKa = 11.29GNDD648 pKa = 2.93ILYY651 pKa = 10.75GDD653 pKa = 5.23DD654 pKa = 4.18GNDD657 pKa = 3.12VIYY660 pKa = 10.63GDD662 pKa = 4.01EE663 pKa = 4.47GADD666 pKa = 3.46VLYY669 pKa = 11.06GGAGNDD675 pKa = 3.51HH676 pKa = 7.3LYY678 pKa = 11.21GGDD681 pKa = 3.79GNDD684 pKa = 4.35TIHH687 pKa = 7.18GEE689 pKa = 4.2YY690 pKa = 11.13GNDD693 pKa = 3.64TLDD696 pKa = 3.84GGAGGNFLSALTGDD710 pKa = 3.62DD711 pKa = 3.39TFVYY715 pKa = 10.47GGGHH719 pKa = 6.38DD720 pKa = 4.25VISDD724 pKa = 3.58TGGSDD729 pKa = 4.02QLLLPSGIVLGDD741 pKa = 3.28LTFQRR746 pKa = 11.84VSTEE750 pKa = 3.67GSLSYY755 pKa = 10.93FDD757 pKa = 5.75DD758 pKa = 4.21LLINIDD764 pKa = 3.53GAGTIEE770 pKa = 4.3IKK772 pKa = 10.84AHH774 pKa = 5.33FQSSSFRR781 pKa = 11.84IEE783 pKa = 3.73TLVFSDD789 pKa = 4.32TSTLDD794 pKa = 3.21LTTLANPDD802 pKa = 3.31VYY804 pKa = 10.47LTAGNDD810 pKa = 3.53TFTSGATGAFAIYY823 pKa = 10.28GGDD826 pKa = 3.43GDD828 pKa = 6.13DD829 pKa = 5.15YY830 pKa = 11.43MSDD833 pKa = 3.86YY834 pKa = 11.39QNAAHH839 pKa = 6.51TFDD842 pKa = 4.47GGNGNDD848 pKa = 3.52AMYY851 pKa = 10.76GGAGNDD857 pKa = 3.48TYY859 pKa = 10.91IASTGFDD866 pKa = 3.91TISEE870 pKa = 4.25NAGTDD875 pKa = 3.83TIVVPAAFTIDD886 pKa = 4.02DD887 pKa = 3.7VTFYY891 pKa = 10.99RR892 pKa = 11.84ILNGSGPTDD901 pKa = 3.31NLGISINGLGEE912 pKa = 3.82IAVIAQFSSALVEE925 pKa = 4.32YY926 pKa = 9.8MHH928 pKa = 7.26FDD930 pKa = 3.48NDD932 pKa = 3.05NSTISLSGLSITTVGTAGNDD952 pKa = 3.45NLSPPSTHH960 pKa = 7.06AGANDD965 pKa = 3.29ILDD968 pKa = 3.73GRR970 pKa = 11.84EE971 pKa = 3.41GDD973 pKa = 4.18DD974 pKa = 3.65YY975 pKa = 12.04VNGGSGDD982 pKa = 3.52DD983 pKa = 3.68TYY985 pKa = 11.31IFSAGHH991 pKa = 5.14DD992 pKa = 4.11TITEE996 pKa = 4.04SSGDD1000 pKa = 3.47DD1001 pKa = 3.69TILVRR1006 pKa = 11.84SSYY1009 pKa = 11.47APGDD1013 pKa = 3.38ISISFVPYY1021 pKa = 10.49YY1022 pKa = 10.94YY1023 pKa = 9.57DD1024 pKa = 3.28TTGMRR1029 pKa = 11.84LEE1031 pKa = 4.51DD1032 pKa = 3.6TDD1034 pKa = 4.19GNSITVYY1041 pKa = 9.82RR1042 pKa = 11.84QGYY1045 pKa = 7.37SSGYY1049 pKa = 8.35VVEE1052 pKa = 5.12HH1053 pKa = 6.15VAFADD1058 pKa = 3.7TTVWNLSSMEE1068 pKa = 4.1IEE1070 pKa = 4.15THH1072 pKa = 5.19GTSSADD1078 pKa = 3.18YY1079 pKa = 10.63LYY1081 pKa = 11.48GHH1083 pKa = 7.34DD1084 pKa = 4.47VGDD1087 pKa = 4.07ASSADD1092 pKa = 3.8TIYY1095 pKa = 10.87GYY1097 pKa = 11.05AGNDD1101 pKa = 3.63TIDD1104 pKa = 3.88AGDD1107 pKa = 4.11GNDD1110 pKa = 4.18LVYY1113 pKa = 11.02AGDD1116 pKa = 3.84GADD1119 pKa = 3.77YY1120 pKa = 11.05VYY1122 pKa = 11.2ASNGNDD1128 pKa = 2.99TVYY1131 pKa = 11.03GGNDD1135 pKa = 3.22ADD1137 pKa = 3.83EE1138 pKa = 4.44LHH1140 pKa = 6.79GKK1142 pKa = 8.37FHH1144 pKa = 6.58SVLYY1148 pKa = 10.94GEE1150 pKa = 5.33AGNDD1154 pKa = 3.27DD1155 pKa = 5.62LYY1157 pKa = 11.73ANPYY1161 pKa = 9.4SNEE1164 pKa = 3.8AAGTLVTLFGGDD1176 pKa = 3.65GADD1179 pKa = 3.45TLHH1182 pKa = 6.51GGNYY1186 pKa = 10.12GEE1188 pKa = 4.53TVMHH1192 pKa = 7.03GGNGVDD1198 pKa = 3.78TMISAYY1204 pKa = 9.35GTDD1207 pKa = 3.31TFAFDD1212 pKa = 3.64AASAFNAVDD1221 pKa = 4.37VIQQFNTSEE1230 pKa = 3.98HH1231 pKa = 6.52DD1232 pKa = 4.06KK1233 pKa = 11.03IDD1235 pKa = 3.42ISDD1238 pKa = 3.97VLDD1241 pKa = 3.47GHH1243 pKa = 6.73YY1244 pKa = 10.96DD1245 pKa = 3.66PLTDD1249 pKa = 5.8AITDD1253 pKa = 4.12FVQITTSGSNSEE1265 pKa = 4.71LFVDD1269 pKa = 4.23TTGSATFGSSQHH1281 pKa = 6.25IATIQYY1287 pKa = 8.05ITGLTDD1293 pKa = 3.17EE1294 pKa = 5.05AALVTAGTLIAAA1306 pKa = 5.1
MM1 pKa = 7.64SGITFIVEE9 pKa = 4.07GATGLDD15 pKa = 3.46EE16 pKa = 4.89QDD18 pKa = 3.39VKK20 pKa = 11.58NSFEE24 pKa = 4.12NLRR27 pKa = 11.84NASGDD32 pKa = 3.37FDD34 pKa = 5.06QDD36 pKa = 2.84ILYY39 pKa = 9.77IASKK43 pKa = 9.3TRR45 pKa = 11.84VLIIIANDD53 pKa = 3.8TVSGTSATYY62 pKa = 9.73PVTSGDD68 pKa = 3.03TAYY71 pKa = 10.96YY72 pKa = 8.45GTYY75 pKa = 9.89DD76 pKa = 3.53YY77 pKa = 11.08VIRR80 pKa = 11.84IPKK83 pKa = 9.84QGFDD87 pKa = 3.3GDD89 pKa = 4.4TYY91 pKa = 11.64NHH93 pKa = 6.52VGTDD97 pKa = 2.81KK98 pKa = 10.76WSLEE102 pKa = 3.53RR103 pKa = 11.84LYY105 pKa = 10.93AHH107 pKa = 6.94EE108 pKa = 4.46VAHH111 pKa = 5.9VMQAIGDD118 pKa = 4.12NAPSTQAEE126 pKa = 4.28RR127 pKa = 11.84EE128 pKa = 4.13EE129 pKa = 4.2QATKK133 pKa = 10.67YY134 pKa = 10.17SNHH137 pKa = 6.11IMSTAHH143 pKa = 6.88PEE145 pKa = 4.03EE146 pKa = 4.88SPQTHH151 pKa = 6.47YY152 pKa = 11.64YY153 pKa = 8.02NTEE156 pKa = 3.76GDD158 pKa = 3.58TGSGLNQNSGSGDD171 pKa = 3.62SPHH174 pKa = 7.97ILDD177 pKa = 4.81DD178 pKa = 4.3SSPLSEE184 pKa = 4.67NLPGAPLPSQLAAIQPAFGDD204 pKa = 4.7AINASSPLVIDD215 pKa = 4.66LSSGHH220 pKa = 6.31SGVTLTTWSASTTEE234 pKa = 4.41TYY236 pKa = 10.87FDD238 pKa = 4.88LDD240 pKa = 4.05DD241 pKa = 4.68NGFAVQTAWVSGDD254 pKa = 3.18TGLLARR260 pKa = 11.84DD261 pKa = 3.68LNEE264 pKa = 3.99NGIIDD269 pKa = 3.83SSAEE273 pKa = 3.95LFGSPTVDD281 pKa = 3.15GFAKK285 pKa = 10.62LAMLDD290 pKa = 3.84SNHH293 pKa = 7.22DD294 pKa = 3.74LRR296 pKa = 11.84IDD298 pKa = 3.96SNDD301 pKa = 3.78DD302 pKa = 2.98AWDD305 pKa = 3.8SLVVWNDD312 pKa = 3.23DD313 pKa = 3.24GDD315 pKa = 4.47AVTQSGEE322 pKa = 4.14LHH324 pKa = 6.37SLASLNIANIDD335 pKa = 3.77LAGVASSTSTISGNSISHH353 pKa = 5.23VSKK356 pKa = 9.69ITFTSGATAAIDD368 pKa = 3.8DD369 pKa = 3.91AWFVHH374 pKa = 7.23DD375 pKa = 4.67NTNSFYY381 pKa = 10.45TGEE384 pKa = 4.13YY385 pKa = 9.05TLDD388 pKa = 3.34VDD390 pKa = 4.06TLFLPRR396 pKa = 11.84VRR398 pKa = 11.84GYY400 pKa = 8.97GTLPDD405 pKa = 3.66LTIAMSLDD413 pKa = 3.57SDD415 pKa = 4.81LKK417 pKa = 11.3DD418 pKa = 3.57LVLGFAGTDD427 pKa = 3.22LSEE430 pKa = 4.23FADD433 pKa = 3.83AKK435 pKa = 10.07TAITDD440 pKa = 4.07ILYY443 pKa = 9.87KK444 pKa = 8.91WAGVEE449 pKa = 4.67GVDD452 pKa = 4.12PDD454 pKa = 4.63SRR456 pKa = 11.84GIWVDD461 pKa = 3.34AQHH464 pKa = 7.13LAFLEE469 pKa = 4.06QFIGSDD475 pKa = 3.48FLQITTHH482 pKa = 6.51SADD485 pKa = 3.58PQPWAGALIEE495 pKa = 4.81DD496 pKa = 5.12AYY498 pKa = 10.94QAAFAMLSADD508 pKa = 3.36ILVQTGFGQLFDD520 pKa = 3.55TAITYY525 pKa = 9.75NPSSGTRR532 pKa = 11.84TGDD535 pKa = 3.14TDD537 pKa = 3.93LSEE540 pKa = 5.05DD541 pKa = 4.59AIDD544 pKa = 4.56DD545 pKa = 4.15LVSIAPSSGPEE556 pKa = 3.71NEE558 pKa = 4.44AFWVSVAWALDD569 pKa = 3.52VTKK572 pKa = 10.81GISNLSYY579 pKa = 11.42DD580 pKa = 3.69EE581 pKa = 4.38TTWLSDD587 pKa = 4.79AITASNALSDD597 pKa = 3.8WATVLHH603 pKa = 6.74SYY605 pKa = 10.64DD606 pKa = 3.81PSAGTASQNGTNGAEE621 pKa = 4.06TLYY624 pKa = 11.42GDD626 pKa = 3.57VTADD630 pKa = 3.87TIHH633 pKa = 6.6GFNGADD639 pKa = 3.82TIYY642 pKa = 11.06GDD644 pKa = 3.8YY645 pKa = 11.29GNDD648 pKa = 2.93ILYY651 pKa = 10.75GDD653 pKa = 5.23DD654 pKa = 4.18GNDD657 pKa = 3.12VIYY660 pKa = 10.63GDD662 pKa = 4.01EE663 pKa = 4.47GADD666 pKa = 3.46VLYY669 pKa = 11.06GGAGNDD675 pKa = 3.51HH676 pKa = 7.3LYY678 pKa = 11.21GGDD681 pKa = 3.79GNDD684 pKa = 4.35TIHH687 pKa = 7.18GEE689 pKa = 4.2YY690 pKa = 11.13GNDD693 pKa = 3.64TLDD696 pKa = 3.84GGAGGNFLSALTGDD710 pKa = 3.62DD711 pKa = 3.39TFVYY715 pKa = 10.47GGGHH719 pKa = 6.38DD720 pKa = 4.25VISDD724 pKa = 3.58TGGSDD729 pKa = 4.02QLLLPSGIVLGDD741 pKa = 3.28LTFQRR746 pKa = 11.84VSTEE750 pKa = 3.67GSLSYY755 pKa = 10.93FDD757 pKa = 5.75DD758 pKa = 4.21LLINIDD764 pKa = 3.53GAGTIEE770 pKa = 4.3IKK772 pKa = 10.84AHH774 pKa = 5.33FQSSSFRR781 pKa = 11.84IEE783 pKa = 3.73TLVFSDD789 pKa = 4.32TSTLDD794 pKa = 3.21LTTLANPDD802 pKa = 3.31VYY804 pKa = 10.47LTAGNDD810 pKa = 3.53TFTSGATGAFAIYY823 pKa = 10.28GGDD826 pKa = 3.43GDD828 pKa = 6.13DD829 pKa = 5.15YY830 pKa = 11.43MSDD833 pKa = 3.86YY834 pKa = 11.39QNAAHH839 pKa = 6.51TFDD842 pKa = 4.47GGNGNDD848 pKa = 3.52AMYY851 pKa = 10.76GGAGNDD857 pKa = 3.48TYY859 pKa = 10.91IASTGFDD866 pKa = 3.91TISEE870 pKa = 4.25NAGTDD875 pKa = 3.83TIVVPAAFTIDD886 pKa = 4.02DD887 pKa = 3.7VTFYY891 pKa = 10.99RR892 pKa = 11.84ILNGSGPTDD901 pKa = 3.31NLGISINGLGEE912 pKa = 3.82IAVIAQFSSALVEE925 pKa = 4.32YY926 pKa = 9.8MHH928 pKa = 7.26FDD930 pKa = 3.48NDD932 pKa = 3.05NSTISLSGLSITTVGTAGNDD952 pKa = 3.45NLSPPSTHH960 pKa = 7.06AGANDD965 pKa = 3.29ILDD968 pKa = 3.73GRR970 pKa = 11.84EE971 pKa = 3.41GDD973 pKa = 4.18DD974 pKa = 3.65YY975 pKa = 12.04VNGGSGDD982 pKa = 3.52DD983 pKa = 3.68TYY985 pKa = 11.31IFSAGHH991 pKa = 5.14DD992 pKa = 4.11TITEE996 pKa = 4.04SSGDD1000 pKa = 3.47DD1001 pKa = 3.69TILVRR1006 pKa = 11.84SSYY1009 pKa = 11.47APGDD1013 pKa = 3.38ISISFVPYY1021 pKa = 10.49YY1022 pKa = 10.94YY1023 pKa = 9.57DD1024 pKa = 3.28TTGMRR1029 pKa = 11.84LEE1031 pKa = 4.51DD1032 pKa = 3.6TDD1034 pKa = 4.19GNSITVYY1041 pKa = 9.82RR1042 pKa = 11.84QGYY1045 pKa = 7.37SSGYY1049 pKa = 8.35VVEE1052 pKa = 5.12HH1053 pKa = 6.15VAFADD1058 pKa = 3.7TTVWNLSSMEE1068 pKa = 4.1IEE1070 pKa = 4.15THH1072 pKa = 5.19GTSSADD1078 pKa = 3.18YY1079 pKa = 10.63LYY1081 pKa = 11.48GHH1083 pKa = 7.34DD1084 pKa = 4.47VGDD1087 pKa = 4.07ASSADD1092 pKa = 3.8TIYY1095 pKa = 10.87GYY1097 pKa = 11.05AGNDD1101 pKa = 3.63TIDD1104 pKa = 3.88AGDD1107 pKa = 4.11GNDD1110 pKa = 4.18LVYY1113 pKa = 11.02AGDD1116 pKa = 3.84GADD1119 pKa = 3.77YY1120 pKa = 11.05VYY1122 pKa = 11.2ASNGNDD1128 pKa = 2.99TVYY1131 pKa = 11.03GGNDD1135 pKa = 3.22ADD1137 pKa = 3.83EE1138 pKa = 4.44LHH1140 pKa = 6.79GKK1142 pKa = 8.37FHH1144 pKa = 6.58SVLYY1148 pKa = 10.94GEE1150 pKa = 5.33AGNDD1154 pKa = 3.27DD1155 pKa = 5.62LYY1157 pKa = 11.73ANPYY1161 pKa = 9.4SNEE1164 pKa = 3.8AAGTLVTLFGGDD1176 pKa = 3.65GADD1179 pKa = 3.45TLHH1182 pKa = 6.51GGNYY1186 pKa = 10.12GEE1188 pKa = 4.53TVMHH1192 pKa = 7.03GGNGVDD1198 pKa = 3.78TMISAYY1204 pKa = 9.35GTDD1207 pKa = 3.31TFAFDD1212 pKa = 3.64AASAFNAVDD1221 pKa = 4.37VIQQFNTSEE1230 pKa = 3.98HH1231 pKa = 6.52DD1232 pKa = 4.06KK1233 pKa = 11.03IDD1235 pKa = 3.42ISDD1238 pKa = 3.97VLDD1241 pKa = 3.47GHH1243 pKa = 6.73YY1244 pKa = 10.96DD1245 pKa = 3.66PLTDD1249 pKa = 5.8AITDD1253 pKa = 4.12FVQITTSGSNSEE1265 pKa = 4.71LFVDD1269 pKa = 4.23TTGSATFGSSQHH1281 pKa = 6.25IATIQYY1287 pKa = 8.05ITGLTDD1293 pKa = 3.17EE1294 pKa = 5.05AALVTAGTLIAAA1306 pKa = 5.1
Molecular weight: 137.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8EPP1|B8EPP1_METSB Multi_ubiq domain-containing protein OS=Methylocella silvestris (strain DSM 15510 / CIP 108128 / LMG 27833 / NCIMB 13906 / BL2) OX=395965 GN=Msil_1952 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84IIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84RR29 pKa = 11.84IIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1219436 |
33 |
2684 |
319.6 |
34.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.821 ± 0.064 | 0.893 ± 0.013 |
5.541 ± 0.032 | 5.588 ± 0.04 |
4.015 ± 0.028 | 8.475 ± 0.037 |
1.888 ± 0.018 | 5.274 ± 0.028 |
3.469 ± 0.032 | 10.231 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.097 ± 0.017 | 2.549 ± 0.024 |
5.481 ± 0.032 | 3.014 ± 0.021 |
7.09 ± 0.043 | 5.636 ± 0.03 |
4.716 ± 0.027 | 6.832 ± 0.034 |
1.222 ± 0.016 | 2.169 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
