
Sulfitobacter indolifex HEL-45
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Sulfitobacter; Sulfitobacter indolifex
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
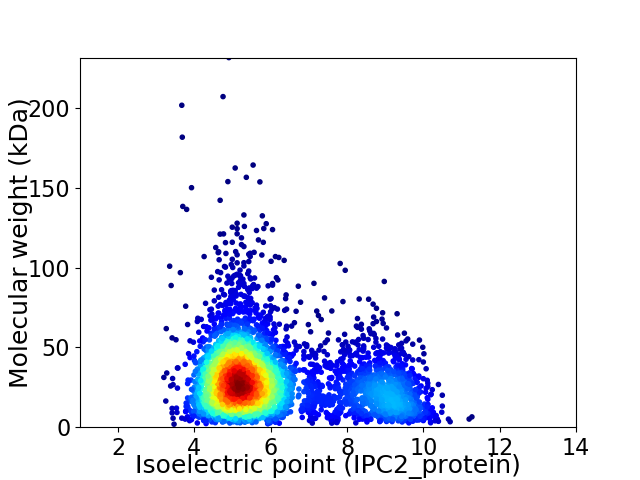
Virtual 2D-PAGE plot for 4147 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A9E7L9|A9E7L9_9RHOB TRAP dicarboxylate transporter-DctP subunit OS=Sulfitobacter indolifex HEL-45 OX=391624 GN=OIHEL45_15224 PE=3 SV=1
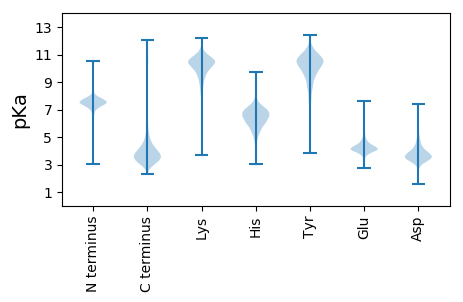
MM1 pKa = 7.35HH2 pKa = 7.4FEE4 pKa = 4.07LSFIGVFLEE13 pKa = 4.64RR14 pKa = 11.84IFGGFFSVARR24 pKa = 11.84RR25 pKa = 11.84GRR27 pKa = 11.84GGAALGLRR35 pKa = 11.84IFCATLLPLVFTTKK49 pKa = 10.54GAFAQQLILNTVGAPTVVGAGIGEE73 pKa = 4.07RR74 pKa = 11.84ALWSNAGTVGGVPVDD89 pKa = 3.83IVAEE93 pKa = 4.2MVFASRR99 pKa = 11.84DD100 pKa = 3.7HH101 pKa = 7.09IFTTGNGQIQITSTGQDD118 pKa = 3.14PHH120 pKa = 8.36FMDD123 pKa = 5.31FYY125 pKa = 10.68LYY127 pKa = 10.63EE128 pKa = 5.35AGTHH132 pKa = 6.64NITTNSGGVPVVADD146 pKa = 3.4VFFQINDD153 pKa = 2.92IDD155 pKa = 4.28GPLNEE160 pKa = 4.01QVYY163 pKa = 10.92VNICDD168 pKa = 3.61GTVEE172 pKa = 3.98YY173 pKa = 11.29VRR175 pKa = 11.84IDD177 pKa = 3.59RR178 pKa = 11.84SATTYY183 pKa = 10.0RR184 pKa = 11.84GYY186 pKa = 10.31IEE188 pKa = 5.93GPDD191 pKa = 3.51ANLGTEE197 pKa = 4.15VFYY200 pKa = 11.04LAGDD204 pKa = 3.81RR205 pKa = 11.84GYY207 pKa = 11.44SNQPVSGLEE216 pKa = 3.33IFYY219 pKa = 9.75PQTSTFNFGRR229 pKa = 11.84TANNGFLVLLSNPTYY244 pKa = 11.1NEE246 pKa = 3.76TQTYY250 pKa = 10.71DD251 pKa = 3.68LQCGDD256 pKa = 4.15FKK258 pKa = 11.69APVLQDD264 pKa = 3.53DD265 pKa = 4.69LKK267 pKa = 10.62EE268 pKa = 4.15QVLSEE273 pKa = 4.03PVVLNILFNDD283 pKa = 3.97SVATEE288 pKa = 4.26NNNAPANNSGEE299 pKa = 3.89PSEE302 pKa = 4.26YY303 pKa = 10.79AKK305 pKa = 10.57QAVDD309 pKa = 5.31LIPPTGALNIVTDD322 pKa = 4.22SEE324 pKa = 4.23GHH326 pKa = 5.37RR327 pKa = 11.84VGFDD331 pKa = 3.18VLGEE335 pKa = 4.39GTWSYY340 pKa = 12.18DD341 pKa = 3.39DD342 pKa = 3.85LTGEE346 pKa = 4.15LTFTPFAAFFAAPTSINYY364 pKa = 9.73RR365 pKa = 11.84YY366 pKa = 9.63QSPIVLPNEE375 pKa = 4.03PQTYY379 pKa = 9.6SAPAQVSIDD388 pKa = 3.71VGSVGLLKK396 pKa = 10.54LAQLVDD402 pKa = 3.88TNLNGYY408 pKa = 9.82ADD410 pKa = 4.18AGEE413 pKa = 4.37TIAYY417 pKa = 9.6VLTAEE422 pKa = 4.28NFGNVDD428 pKa = 3.57LTNVQLAEE436 pKa = 4.18TQFSGKK442 pKa = 9.74GVPPVITFQAATSLSPEE459 pKa = 3.84GTLLVGEE466 pKa = 4.4KK467 pKa = 10.26AVYY470 pKa = 7.91TATYY474 pKa = 9.11TLVPEE479 pKa = 5.2DD480 pKa = 4.76LDD482 pKa = 4.03TTISNQAEE490 pKa = 4.36VTAQTPGGTVVSDD503 pKa = 4.49LSDD506 pKa = 3.56SEE508 pKa = 4.51NPGDD512 pKa = 4.87GDD514 pKa = 3.9GTPSNGPGAGRR525 pKa = 11.84DD526 pKa = 3.85DD527 pKa = 3.86PTTIYY532 pKa = 10.61AGSGPDD538 pKa = 4.01RR539 pKa = 11.84GDD541 pKa = 3.35APITYY546 pKa = 10.05GDD548 pKa = 4.06PQHH551 pKa = 7.13ADD553 pKa = 2.92TSQYY557 pKa = 10.25WIGALNGDD565 pKa = 3.97GDD567 pKa = 4.06GSAQHH572 pKa = 6.5SVDD575 pKa = 3.33GTGDD579 pKa = 3.8DD580 pKa = 5.05LDD582 pKa = 5.42GNDD585 pKa = 5.05DD586 pKa = 3.68EE587 pKa = 5.95SEE589 pKa = 4.01EE590 pKa = 4.82AFPQLYY596 pKa = 10.93GDD598 pKa = 4.44LTRR601 pKa = 11.84TATVVVNEE609 pKa = 4.27PSPGSGYY616 pKa = 10.54LQAFVDD622 pKa = 4.41FGGDD626 pKa = 3.45GLFLGDD632 pKa = 4.83QVATDD637 pKa = 3.73IQDD640 pKa = 3.95GGPQDD645 pKa = 4.7LDD647 pKa = 3.5GAVNGEE653 pKa = 3.67ISFPISVPATAVLTPTFVRR672 pKa = 11.84LRR674 pKa = 11.84WSSAAGLDD682 pKa = 3.83ALTAAPDD689 pKa = 4.19GEE691 pKa = 4.38VEE693 pKa = 5.09DD694 pKa = 4.18YY695 pKa = 11.27GITIKK700 pKa = 10.04TPPDD704 pKa = 3.72ADD706 pKa = 4.12RR707 pKa = 11.84GDD709 pKa = 4.02APASYY714 pKa = 10.82GDD716 pKa = 3.67PQHH719 pKa = 7.62IIEE722 pKa = 5.07GPGAPEE728 pKa = 4.28VYY730 pKa = 10.25LGTVPPDD737 pKa = 3.23IDD739 pKa = 5.23LLAQNSIGATGDD751 pKa = 3.73DD752 pKa = 4.11ADD754 pKa = 5.2GSDD757 pKa = 4.75DD758 pKa = 3.66EE759 pKa = 6.33DD760 pKa = 4.66GVVLPQFYY768 pKa = 10.39RR769 pKa = 11.84GGLAEE774 pKa = 4.28IAVTVNDD781 pKa = 3.55RR782 pKa = 11.84SGAPRR787 pKa = 11.84SSYY790 pKa = 10.63LQAFIDD796 pKa = 4.1FNGDD800 pKa = 3.16GTFGQAGEE808 pKa = 4.2QVALNLQDD816 pKa = 4.29GDD818 pKa = 5.38AIDD821 pKa = 5.3KK822 pKa = 11.09DD823 pKa = 3.91GATNGSILFEE833 pKa = 4.24VAVPAGATTLPTYY846 pKa = 11.12ARR848 pKa = 11.84FRR850 pKa = 11.84WSTDD854 pKa = 2.97SAGLATAFEE863 pKa = 5.05GEE865 pKa = 4.33VEE867 pKa = 5.13DD868 pKa = 3.89YY869 pKa = 10.61TLTISSDD876 pKa = 3.91PPPLVCDD883 pKa = 3.85ASLYY887 pKa = 10.84AIVDD891 pKa = 3.92SPSTLIRR898 pKa = 11.84MTFRR902 pKa = 11.84DD903 pKa = 3.47AGGSYY908 pKa = 9.14ATTVDD913 pKa = 3.66PSGSTGEE920 pKa = 4.24NRR922 pKa = 11.84DD923 pKa = 3.6GAWGYY928 pKa = 10.32NALDD932 pKa = 4.1GYY934 pKa = 10.72IYY936 pKa = 10.57GVEE939 pKa = 3.99EE940 pKa = 3.77NTRR943 pKa = 11.84RR944 pKa = 11.84LYY946 pKa = 10.84RR947 pKa = 11.84IDD949 pKa = 3.45GAGNIADD956 pKa = 4.71LGNISGADD964 pKa = 3.52DD965 pKa = 4.22ALNAGDD971 pKa = 4.32ILPNGRR977 pKa = 11.84FIYY980 pKa = 10.37EE981 pKa = 3.52VDD983 pKa = 3.21ANTWQIVDD991 pKa = 4.35LTNPTAPIGLGAIALTIGVNPEE1013 pKa = 3.77DD1014 pKa = 4.06LAFNPVDD1021 pKa = 3.99GIMYY1025 pKa = 10.39GIDD1028 pKa = 3.37RR1029 pKa = 11.84NSGFLFRR1036 pKa = 11.84AAVNNGTPGSVTPQAIGPAVYY1057 pKa = 10.59AGMFDD1062 pKa = 4.52ALWFDD1067 pKa = 3.49RR1068 pKa = 11.84DD1069 pKa = 3.31GRR1071 pKa = 11.84LYY1073 pKa = 10.86GYY1075 pKa = 11.32SNTTNNLFIISTATGVPRR1093 pKa = 11.84LISNIPGDD1101 pKa = 3.69EE1102 pKa = 4.08GDD1104 pKa = 3.71RR1105 pKa = 11.84SDD1107 pKa = 5.64GISCRR1112 pKa = 11.84GPAPIPLGGISGNIYY1127 pKa = 10.52EE1128 pKa = 5.25DD1129 pKa = 4.07ADD1131 pKa = 3.75ASDD1134 pKa = 4.1VKK1136 pKa = 11.24DD1137 pKa = 3.46SGEE1140 pKa = 4.34TNFGAGVGISVYY1152 pKa = 11.05ADD1154 pKa = 2.85NGTPANTADD1163 pKa = 4.32DD1164 pKa = 4.28VFLVTTDD1171 pKa = 3.31TLADD1175 pKa = 3.31GTYY1178 pKa = 10.96AVGDD1182 pKa = 3.63LMVNTTYY1189 pKa = 10.74RR1190 pKa = 11.84VALDD1194 pKa = 3.86EE1195 pKa = 5.16ADD1197 pKa = 3.97ADD1199 pKa = 4.25LPSGLTIGTSNPLIGVAVSGNSVTTDD1225 pKa = 2.75QDD1227 pKa = 3.61FGFDD1231 pKa = 3.63PAGSDD1236 pKa = 4.64LEE1238 pKa = 4.32LTKK1241 pKa = 10.35IAAATGTITPITNVSEE1257 pKa = 4.16GDD1259 pKa = 3.63TIDD1262 pKa = 3.63WIITITNVSGGSPSGVKK1279 pKa = 10.51VIDD1282 pKa = 3.91QIPSGFAYY1290 pKa = 10.69VSDD1293 pKa = 4.76DD1294 pKa = 3.83APSTGDD1300 pKa = 3.23TYY1302 pKa = 11.75DD1303 pKa = 4.03PNTGLWFVDD1312 pKa = 3.74EE1313 pKa = 4.7LLSGASEE1320 pKa = 4.24TLTITTTVLGSGDD1333 pKa = 3.63FTNQAEE1339 pKa = 4.73IIYY1342 pKa = 10.38SSLPDD1347 pKa = 4.47PDD1349 pKa = 4.77SDD1351 pKa = 4.64PNTGPLTDD1359 pKa = 5.22DD1360 pKa = 5.62LFDD1363 pKa = 4.06QLPDD1367 pKa = 4.15DD1368 pKa = 5.46DD1369 pKa = 4.34EE1370 pKa = 6.53ASYY1373 pKa = 11.37SVNLVTGEE1381 pKa = 4.38RR1382 pKa = 11.84ILSGRR1387 pKa = 11.84VFIDD1391 pKa = 3.0NGAGGGTAHH1400 pKa = 7.69DD1401 pKa = 4.06ALRR1404 pKa = 11.84NGSEE1408 pKa = 4.2SGASSAVFEE1417 pKa = 4.76ILDD1420 pKa = 3.47SSGTVLATPGVAADD1434 pKa = 4.15GTWSYY1439 pKa = 11.21GLSGTYY1445 pKa = 10.01SGAITIRR1452 pKa = 11.84AIPANDD1458 pKa = 3.03YY1459 pKa = 10.66RR1460 pKa = 11.84AISEE1464 pKa = 4.33ATSGLPDD1471 pKa = 3.82LVDD1474 pKa = 4.61ANPHH1478 pKa = 6.18DD1479 pKa = 4.45GEE1481 pKa = 4.53FTFTPEE1487 pKa = 3.77AFGNRR1492 pKa = 11.84MGLDD1496 pKa = 2.87IGLLQLPSLTNDD1508 pKa = 2.77QTTTVAQGQVATLPHH1523 pKa = 7.47IYY1525 pKa = 9.14TATSEE1530 pKa = 4.38GQVTFSFAGATSTPTGAFNAALYY1553 pKa = 10.44QDD1555 pKa = 4.24VGCDD1559 pKa = 3.61DD1560 pKa = 4.81SLDD1563 pKa = 3.89GPITGPIAVITGQTICIVSRR1583 pKa = 11.84VSAGSGAGQGSTYY1596 pKa = 10.5VYY1598 pKa = 10.45QVLAATAFARR1608 pKa = 11.84TTVTSSVSNTDD1619 pKa = 3.6EE1620 pKa = 4.28ISVGGQSTQIEE1631 pKa = 4.17LRR1633 pKa = 11.84KK1634 pKa = 7.86TVEE1637 pKa = 4.02NEE1639 pKa = 3.98TQGTLEE1645 pKa = 3.98GTTNLGSINDD1655 pKa = 3.47ILLYY1659 pKa = 10.35RR1660 pKa = 11.84IYY1662 pKa = 10.72LRR1664 pKa = 11.84NKK1666 pKa = 9.05ATTQASNVKK1675 pKa = 9.32IYY1677 pKa = 11.35DD1678 pKa = 3.28MTPPYY1683 pKa = 10.04TSLSEE1688 pKa = 4.64PIVDD1692 pKa = 3.85PQQVSPNLNCDD1703 pKa = 3.64LVRR1706 pKa = 11.84PASNVASYY1714 pKa = 11.18AGAIEE1719 pKa = 4.35WNCTGQMLPGEE1730 pKa = 4.3TGAVQFRR1737 pKa = 11.84VGIQQ1741 pKa = 2.88
MM1 pKa = 7.35HH2 pKa = 7.4FEE4 pKa = 4.07LSFIGVFLEE13 pKa = 4.64RR14 pKa = 11.84IFGGFFSVARR24 pKa = 11.84RR25 pKa = 11.84GRR27 pKa = 11.84GGAALGLRR35 pKa = 11.84IFCATLLPLVFTTKK49 pKa = 10.54GAFAQQLILNTVGAPTVVGAGIGEE73 pKa = 4.07RR74 pKa = 11.84ALWSNAGTVGGVPVDD89 pKa = 3.83IVAEE93 pKa = 4.2MVFASRR99 pKa = 11.84DD100 pKa = 3.7HH101 pKa = 7.09IFTTGNGQIQITSTGQDD118 pKa = 3.14PHH120 pKa = 8.36FMDD123 pKa = 5.31FYY125 pKa = 10.68LYY127 pKa = 10.63EE128 pKa = 5.35AGTHH132 pKa = 6.64NITTNSGGVPVVADD146 pKa = 3.4VFFQINDD153 pKa = 2.92IDD155 pKa = 4.28GPLNEE160 pKa = 4.01QVYY163 pKa = 10.92VNICDD168 pKa = 3.61GTVEE172 pKa = 3.98YY173 pKa = 11.29VRR175 pKa = 11.84IDD177 pKa = 3.59RR178 pKa = 11.84SATTYY183 pKa = 10.0RR184 pKa = 11.84GYY186 pKa = 10.31IEE188 pKa = 5.93GPDD191 pKa = 3.51ANLGTEE197 pKa = 4.15VFYY200 pKa = 11.04LAGDD204 pKa = 3.81RR205 pKa = 11.84GYY207 pKa = 11.44SNQPVSGLEE216 pKa = 3.33IFYY219 pKa = 9.75PQTSTFNFGRR229 pKa = 11.84TANNGFLVLLSNPTYY244 pKa = 11.1NEE246 pKa = 3.76TQTYY250 pKa = 10.71DD251 pKa = 3.68LQCGDD256 pKa = 4.15FKK258 pKa = 11.69APVLQDD264 pKa = 3.53DD265 pKa = 4.69LKK267 pKa = 10.62EE268 pKa = 4.15QVLSEE273 pKa = 4.03PVVLNILFNDD283 pKa = 3.97SVATEE288 pKa = 4.26NNNAPANNSGEE299 pKa = 3.89PSEE302 pKa = 4.26YY303 pKa = 10.79AKK305 pKa = 10.57QAVDD309 pKa = 5.31LIPPTGALNIVTDD322 pKa = 4.22SEE324 pKa = 4.23GHH326 pKa = 5.37RR327 pKa = 11.84VGFDD331 pKa = 3.18VLGEE335 pKa = 4.39GTWSYY340 pKa = 12.18DD341 pKa = 3.39DD342 pKa = 3.85LTGEE346 pKa = 4.15LTFTPFAAFFAAPTSINYY364 pKa = 9.73RR365 pKa = 11.84YY366 pKa = 9.63QSPIVLPNEE375 pKa = 4.03PQTYY379 pKa = 9.6SAPAQVSIDD388 pKa = 3.71VGSVGLLKK396 pKa = 10.54LAQLVDD402 pKa = 3.88TNLNGYY408 pKa = 9.82ADD410 pKa = 4.18AGEE413 pKa = 4.37TIAYY417 pKa = 9.6VLTAEE422 pKa = 4.28NFGNVDD428 pKa = 3.57LTNVQLAEE436 pKa = 4.18TQFSGKK442 pKa = 9.74GVPPVITFQAATSLSPEE459 pKa = 3.84GTLLVGEE466 pKa = 4.4KK467 pKa = 10.26AVYY470 pKa = 7.91TATYY474 pKa = 9.11TLVPEE479 pKa = 5.2DD480 pKa = 4.76LDD482 pKa = 4.03TTISNQAEE490 pKa = 4.36VTAQTPGGTVVSDD503 pKa = 4.49LSDD506 pKa = 3.56SEE508 pKa = 4.51NPGDD512 pKa = 4.87GDD514 pKa = 3.9GTPSNGPGAGRR525 pKa = 11.84DD526 pKa = 3.85DD527 pKa = 3.86PTTIYY532 pKa = 10.61AGSGPDD538 pKa = 4.01RR539 pKa = 11.84GDD541 pKa = 3.35APITYY546 pKa = 10.05GDD548 pKa = 4.06PQHH551 pKa = 7.13ADD553 pKa = 2.92TSQYY557 pKa = 10.25WIGALNGDD565 pKa = 3.97GDD567 pKa = 4.06GSAQHH572 pKa = 6.5SVDD575 pKa = 3.33GTGDD579 pKa = 3.8DD580 pKa = 5.05LDD582 pKa = 5.42GNDD585 pKa = 5.05DD586 pKa = 3.68EE587 pKa = 5.95SEE589 pKa = 4.01EE590 pKa = 4.82AFPQLYY596 pKa = 10.93GDD598 pKa = 4.44LTRR601 pKa = 11.84TATVVVNEE609 pKa = 4.27PSPGSGYY616 pKa = 10.54LQAFVDD622 pKa = 4.41FGGDD626 pKa = 3.45GLFLGDD632 pKa = 4.83QVATDD637 pKa = 3.73IQDD640 pKa = 3.95GGPQDD645 pKa = 4.7LDD647 pKa = 3.5GAVNGEE653 pKa = 3.67ISFPISVPATAVLTPTFVRR672 pKa = 11.84LRR674 pKa = 11.84WSSAAGLDD682 pKa = 3.83ALTAAPDD689 pKa = 4.19GEE691 pKa = 4.38VEE693 pKa = 5.09DD694 pKa = 4.18YY695 pKa = 11.27GITIKK700 pKa = 10.04TPPDD704 pKa = 3.72ADD706 pKa = 4.12RR707 pKa = 11.84GDD709 pKa = 4.02APASYY714 pKa = 10.82GDD716 pKa = 3.67PQHH719 pKa = 7.62IIEE722 pKa = 5.07GPGAPEE728 pKa = 4.28VYY730 pKa = 10.25LGTVPPDD737 pKa = 3.23IDD739 pKa = 5.23LLAQNSIGATGDD751 pKa = 3.73DD752 pKa = 4.11ADD754 pKa = 5.2GSDD757 pKa = 4.75DD758 pKa = 3.66EE759 pKa = 6.33DD760 pKa = 4.66GVVLPQFYY768 pKa = 10.39RR769 pKa = 11.84GGLAEE774 pKa = 4.28IAVTVNDD781 pKa = 3.55RR782 pKa = 11.84SGAPRR787 pKa = 11.84SSYY790 pKa = 10.63LQAFIDD796 pKa = 4.1FNGDD800 pKa = 3.16GTFGQAGEE808 pKa = 4.2QVALNLQDD816 pKa = 4.29GDD818 pKa = 5.38AIDD821 pKa = 5.3KK822 pKa = 11.09DD823 pKa = 3.91GATNGSILFEE833 pKa = 4.24VAVPAGATTLPTYY846 pKa = 11.12ARR848 pKa = 11.84FRR850 pKa = 11.84WSTDD854 pKa = 2.97SAGLATAFEE863 pKa = 5.05GEE865 pKa = 4.33VEE867 pKa = 5.13DD868 pKa = 3.89YY869 pKa = 10.61TLTISSDD876 pKa = 3.91PPPLVCDD883 pKa = 3.85ASLYY887 pKa = 10.84AIVDD891 pKa = 3.92SPSTLIRR898 pKa = 11.84MTFRR902 pKa = 11.84DD903 pKa = 3.47AGGSYY908 pKa = 9.14ATTVDD913 pKa = 3.66PSGSTGEE920 pKa = 4.24NRR922 pKa = 11.84DD923 pKa = 3.6GAWGYY928 pKa = 10.32NALDD932 pKa = 4.1GYY934 pKa = 10.72IYY936 pKa = 10.57GVEE939 pKa = 3.99EE940 pKa = 3.77NTRR943 pKa = 11.84RR944 pKa = 11.84LYY946 pKa = 10.84RR947 pKa = 11.84IDD949 pKa = 3.45GAGNIADD956 pKa = 4.71LGNISGADD964 pKa = 3.52DD965 pKa = 4.22ALNAGDD971 pKa = 4.32ILPNGRR977 pKa = 11.84FIYY980 pKa = 10.37EE981 pKa = 3.52VDD983 pKa = 3.21ANTWQIVDD991 pKa = 4.35LTNPTAPIGLGAIALTIGVNPEE1013 pKa = 3.77DD1014 pKa = 4.06LAFNPVDD1021 pKa = 3.99GIMYY1025 pKa = 10.39GIDD1028 pKa = 3.37RR1029 pKa = 11.84NSGFLFRR1036 pKa = 11.84AAVNNGTPGSVTPQAIGPAVYY1057 pKa = 10.59AGMFDD1062 pKa = 4.52ALWFDD1067 pKa = 3.49RR1068 pKa = 11.84DD1069 pKa = 3.31GRR1071 pKa = 11.84LYY1073 pKa = 10.86GYY1075 pKa = 11.32SNTTNNLFIISTATGVPRR1093 pKa = 11.84LISNIPGDD1101 pKa = 3.69EE1102 pKa = 4.08GDD1104 pKa = 3.71RR1105 pKa = 11.84SDD1107 pKa = 5.64GISCRR1112 pKa = 11.84GPAPIPLGGISGNIYY1127 pKa = 10.52EE1128 pKa = 5.25DD1129 pKa = 4.07ADD1131 pKa = 3.75ASDD1134 pKa = 4.1VKK1136 pKa = 11.24DD1137 pKa = 3.46SGEE1140 pKa = 4.34TNFGAGVGISVYY1152 pKa = 11.05ADD1154 pKa = 2.85NGTPANTADD1163 pKa = 4.32DD1164 pKa = 4.28VFLVTTDD1171 pKa = 3.31TLADD1175 pKa = 3.31GTYY1178 pKa = 10.96AVGDD1182 pKa = 3.63LMVNTTYY1189 pKa = 10.74RR1190 pKa = 11.84VALDD1194 pKa = 3.86EE1195 pKa = 5.16ADD1197 pKa = 3.97ADD1199 pKa = 4.25LPSGLTIGTSNPLIGVAVSGNSVTTDD1225 pKa = 2.75QDD1227 pKa = 3.61FGFDD1231 pKa = 3.63PAGSDD1236 pKa = 4.64LEE1238 pKa = 4.32LTKK1241 pKa = 10.35IAAATGTITPITNVSEE1257 pKa = 4.16GDD1259 pKa = 3.63TIDD1262 pKa = 3.63WIITITNVSGGSPSGVKK1279 pKa = 10.51VIDD1282 pKa = 3.91QIPSGFAYY1290 pKa = 10.69VSDD1293 pKa = 4.76DD1294 pKa = 3.83APSTGDD1300 pKa = 3.23TYY1302 pKa = 11.75DD1303 pKa = 4.03PNTGLWFVDD1312 pKa = 3.74EE1313 pKa = 4.7LLSGASEE1320 pKa = 4.24TLTITTTVLGSGDD1333 pKa = 3.63FTNQAEE1339 pKa = 4.73IIYY1342 pKa = 10.38SSLPDD1347 pKa = 4.47PDD1349 pKa = 4.77SDD1351 pKa = 4.64PNTGPLTDD1359 pKa = 5.22DD1360 pKa = 5.62LFDD1363 pKa = 4.06QLPDD1367 pKa = 4.15DD1368 pKa = 5.46DD1369 pKa = 4.34EE1370 pKa = 6.53ASYY1373 pKa = 11.37SVNLVTGEE1381 pKa = 4.38RR1382 pKa = 11.84ILSGRR1387 pKa = 11.84VFIDD1391 pKa = 3.0NGAGGGTAHH1400 pKa = 7.69DD1401 pKa = 4.06ALRR1404 pKa = 11.84NGSEE1408 pKa = 4.2SGASSAVFEE1417 pKa = 4.76ILDD1420 pKa = 3.47SSGTVLATPGVAADD1434 pKa = 4.15GTWSYY1439 pKa = 11.21GLSGTYY1445 pKa = 10.01SGAITIRR1452 pKa = 11.84AIPANDD1458 pKa = 3.03YY1459 pKa = 10.66RR1460 pKa = 11.84AISEE1464 pKa = 4.33ATSGLPDD1471 pKa = 3.82LVDD1474 pKa = 4.61ANPHH1478 pKa = 6.18DD1479 pKa = 4.45GEE1481 pKa = 4.53FTFTPEE1487 pKa = 3.77AFGNRR1492 pKa = 11.84MGLDD1496 pKa = 2.87IGLLQLPSLTNDD1508 pKa = 2.77QTTTVAQGQVATLPHH1523 pKa = 7.47IYY1525 pKa = 9.14TATSEE1530 pKa = 4.38GQVTFSFAGATSTPTGAFNAALYY1553 pKa = 10.44QDD1555 pKa = 4.24VGCDD1559 pKa = 3.61DD1560 pKa = 4.81SLDD1563 pKa = 3.89GPITGPIAVITGQTICIVSRR1583 pKa = 11.84VSAGSGAGQGSTYY1596 pKa = 10.5VYY1598 pKa = 10.45QVLAATAFARR1608 pKa = 11.84TTVTSSVSNTDD1619 pKa = 3.6EE1620 pKa = 4.28ISVGGQSTQIEE1631 pKa = 4.17LRR1633 pKa = 11.84KK1634 pKa = 7.86TVEE1637 pKa = 4.02NEE1639 pKa = 3.98TQGTLEE1645 pKa = 3.98GTTNLGSINDD1655 pKa = 3.47ILLYY1659 pKa = 10.35RR1660 pKa = 11.84IYY1662 pKa = 10.72LRR1664 pKa = 11.84NKK1666 pKa = 9.05ATTQASNVKK1675 pKa = 9.32IYY1677 pKa = 11.35DD1678 pKa = 3.28MTPPYY1683 pKa = 10.04TSLSEE1688 pKa = 4.64PIVDD1692 pKa = 3.85PQQVSPNLNCDD1703 pKa = 3.64LVRR1706 pKa = 11.84PASNVASYY1714 pKa = 11.18AGAIEE1719 pKa = 4.35WNCTGQMLPGEE1730 pKa = 4.3TGAVQFRR1737 pKa = 11.84VGIQQ1741 pKa = 2.88
Molecular weight: 181.81 kDa
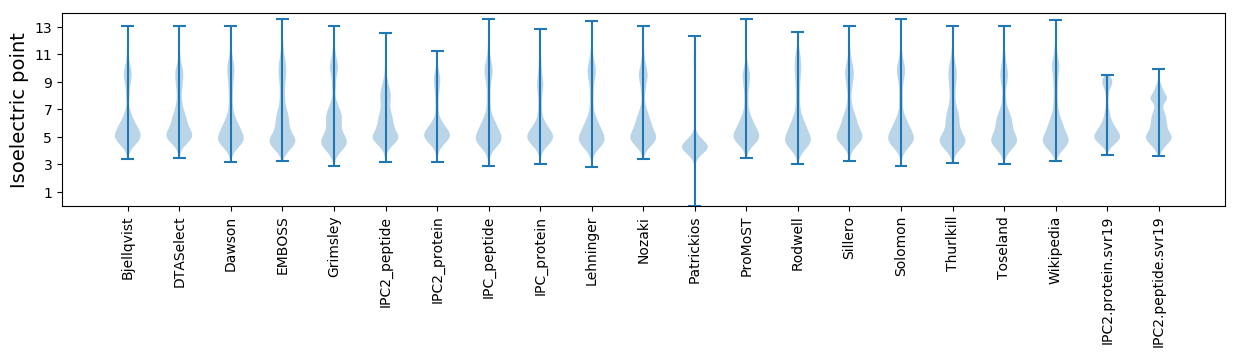
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A9E5V9|A9E5V9_9RHOB Cysteine--tRNA ligase OS=Sulfitobacter indolifex HEL-45 OX=391624 GN=cysS PE=3 SV=1
MM1 pKa = 7.23NSIVNMVMRR10 pKa = 11.84QVMRR14 pKa = 11.84RR15 pKa = 11.84LVSRR19 pKa = 11.84GIGAGTRR26 pKa = 11.84RR27 pKa = 11.84AGKK30 pKa = 9.92RR31 pKa = 11.84GSGKK35 pKa = 10.46GGAFNARR42 pKa = 11.84TAQQGIRR49 pKa = 11.84LLRR52 pKa = 11.84RR53 pKa = 11.84FFRR56 pKa = 11.84SKK58 pKa = 10.99
MM1 pKa = 7.23NSIVNMVMRR10 pKa = 11.84QVMRR14 pKa = 11.84RR15 pKa = 11.84LVSRR19 pKa = 11.84GIGAGTRR26 pKa = 11.84RR27 pKa = 11.84AGKK30 pKa = 9.92RR31 pKa = 11.84GSGKK35 pKa = 10.46GGAFNARR42 pKa = 11.84TAQQGIRR49 pKa = 11.84LLRR52 pKa = 11.84RR53 pKa = 11.84FFRR56 pKa = 11.84SKK58 pKa = 10.99
Molecular weight: 6.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1216087 |
20 |
2148 |
293.2 |
31.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.224 ± 0.055 | 0.89 ± 0.011 |
6.095 ± 0.034 | 5.987 ± 0.039 |
3.693 ± 0.024 | 8.533 ± 0.039 |
2.07 ± 0.021 | 5.223 ± 0.027 |
3.297 ± 0.031 | 10.07 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.898 ± 0.019 | 2.709 ± 0.019 |
4.971 ± 0.024 | 3.369 ± 0.021 |
6.466 ± 0.037 | 5.199 ± 0.025 |
5.548 ± 0.027 | 7.185 ± 0.032 |
1.349 ± 0.017 | 2.221 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
