
Munguba virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Munguba phlebovirus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
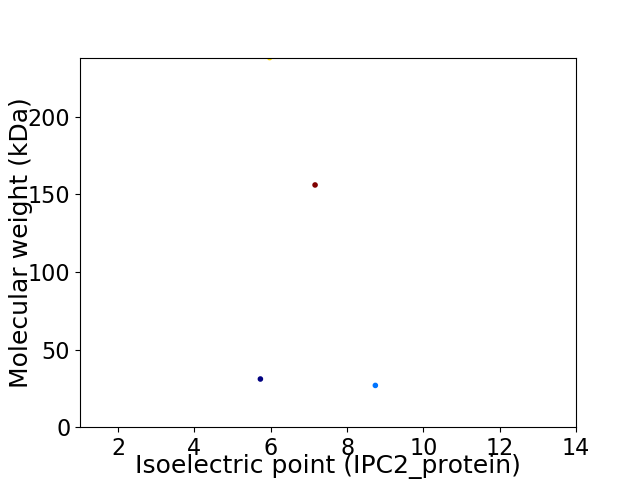
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1T343|I1T343_9VIRU Nonstructural protein OS=Munguba virus OX=1048854 PE=4 SV=1
MM1 pKa = 7.37SFLWNLPVVRR11 pKa = 11.84PSSCFLGLPSVNYY24 pKa = 10.0VAFNKK29 pKa = 9.24MATFEE34 pKa = 4.11ACKK37 pKa = 10.79VNGAEE42 pKa = 5.15IPLKK46 pKa = 10.34HH47 pKa = 6.8LKK49 pKa = 10.06LCYY52 pKa = 9.94DD53 pKa = 3.49KK54 pKa = 11.22RR55 pKa = 11.84PTLFDD60 pKa = 3.49FLRR63 pKa = 11.84NRR65 pKa = 11.84EE66 pKa = 4.2VPVSWGDD73 pKa = 3.48HH74 pKa = 5.87TDD76 pKa = 3.54DD77 pKa = 4.74QILNRR82 pKa = 11.84SPRR85 pKa = 11.84TFEE88 pKa = 3.79GLMFGLSEE96 pKa = 4.14IEE98 pKa = 3.79EE99 pKa = 4.39VEE101 pKa = 3.92YY102 pKa = 11.2GRR104 pKa = 11.84YY105 pKa = 9.02NEE107 pKa = 4.8PYY109 pKa = 9.96LKK111 pKa = 10.55KK112 pKa = 10.46AVSWPLSYY120 pKa = 10.31PSHH123 pKa = 6.11TFFNLAVRR131 pKa = 11.84KK132 pKa = 8.16TEE134 pKa = 3.75MGNWLHH140 pKa = 7.04SSMWATMMLRR150 pKa = 11.84MSHH153 pKa = 6.01TDD155 pKa = 3.12NLEE158 pKa = 3.52EE159 pKa = 4.96SIIKK163 pKa = 8.91VRR165 pKa = 11.84KK166 pKa = 9.81RR167 pKa = 11.84IIEE170 pKa = 4.05KK171 pKa = 10.71ANDD174 pKa = 3.32MGLDD178 pKa = 3.7LSLFTGEE185 pKa = 4.39NLILEE190 pKa = 4.57IAHH193 pKa = 6.04LQCLRR198 pKa = 11.84LIYY201 pKa = 10.32ASRR204 pKa = 11.84KK205 pKa = 6.85EE206 pKa = 3.87RR207 pKa = 11.84EE208 pKa = 3.77RR209 pKa = 11.84GVNHH213 pKa = 6.65SKK215 pKa = 9.48TYY217 pKa = 10.4SIVRR221 pKa = 11.84SYY223 pKa = 11.89DD224 pKa = 3.47CFSVISDD231 pKa = 3.65VNFDD235 pKa = 3.72PMIADD240 pKa = 3.87SLFGSNDD247 pKa = 2.61GTMSGEE253 pKa = 3.97SDD255 pKa = 3.18VDD257 pKa = 3.41QLRR260 pKa = 11.84VDD262 pKa = 3.92LEE264 pKa = 4.66RR265 pKa = 11.84YY266 pKa = 9.71LSLL269 pKa = 5.29
MM1 pKa = 7.37SFLWNLPVVRR11 pKa = 11.84PSSCFLGLPSVNYY24 pKa = 10.0VAFNKK29 pKa = 9.24MATFEE34 pKa = 4.11ACKK37 pKa = 10.79VNGAEE42 pKa = 5.15IPLKK46 pKa = 10.34HH47 pKa = 6.8LKK49 pKa = 10.06LCYY52 pKa = 9.94DD53 pKa = 3.49KK54 pKa = 11.22RR55 pKa = 11.84PTLFDD60 pKa = 3.49FLRR63 pKa = 11.84NRR65 pKa = 11.84EE66 pKa = 4.2VPVSWGDD73 pKa = 3.48HH74 pKa = 5.87TDD76 pKa = 3.54DD77 pKa = 4.74QILNRR82 pKa = 11.84SPRR85 pKa = 11.84TFEE88 pKa = 3.79GLMFGLSEE96 pKa = 4.14IEE98 pKa = 3.79EE99 pKa = 4.39VEE101 pKa = 3.92YY102 pKa = 11.2GRR104 pKa = 11.84YY105 pKa = 9.02NEE107 pKa = 4.8PYY109 pKa = 9.96LKK111 pKa = 10.55KK112 pKa = 10.46AVSWPLSYY120 pKa = 10.31PSHH123 pKa = 6.11TFFNLAVRR131 pKa = 11.84KK132 pKa = 8.16TEE134 pKa = 3.75MGNWLHH140 pKa = 7.04SSMWATMMLRR150 pKa = 11.84MSHH153 pKa = 6.01TDD155 pKa = 3.12NLEE158 pKa = 3.52EE159 pKa = 4.96SIIKK163 pKa = 8.91VRR165 pKa = 11.84KK166 pKa = 9.81RR167 pKa = 11.84IIEE170 pKa = 4.05KK171 pKa = 10.71ANDD174 pKa = 3.32MGLDD178 pKa = 3.7LSLFTGEE185 pKa = 4.39NLILEE190 pKa = 4.57IAHH193 pKa = 6.04LQCLRR198 pKa = 11.84LIYY201 pKa = 10.32ASRR204 pKa = 11.84KK205 pKa = 6.85EE206 pKa = 3.87RR207 pKa = 11.84EE208 pKa = 3.77RR209 pKa = 11.84GVNHH213 pKa = 6.65SKK215 pKa = 9.48TYY217 pKa = 10.4SIVRR221 pKa = 11.84SYY223 pKa = 11.89DD224 pKa = 3.47CFSVISDD231 pKa = 3.65VNFDD235 pKa = 3.72PMIADD240 pKa = 3.87SLFGSNDD247 pKa = 2.61GTMSGEE253 pKa = 3.97SDD255 pKa = 3.18VDD257 pKa = 3.41QLRR260 pKa = 11.84VDD262 pKa = 3.92LEE264 pKa = 4.66RR265 pKa = 11.84YY266 pKa = 9.71LSLL269 pKa = 5.29
Molecular weight: 31.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1T343|I1T343_9VIRU Nonstructural protein OS=Munguba virus OX=1048854 PE=4 SV=1
MM1 pKa = 7.2SADD4 pKa = 3.58YY5 pKa = 10.81QRR7 pKa = 11.84IAIEE11 pKa = 4.55FGSEE15 pKa = 3.99PIDD18 pKa = 3.8RR19 pKa = 11.84DD20 pKa = 5.19AITLWVNEE28 pKa = 3.91FAYY31 pKa = 10.56QGFDD35 pKa = 2.87ASNVLRR41 pKa = 11.84LLQQRR46 pKa = 11.84GGATWKK52 pKa = 10.48EE53 pKa = 4.07DD54 pKa = 3.36AKK56 pKa = 11.53KK57 pKa = 10.17MIILVLTRR65 pKa = 11.84GNKK68 pKa = 8.28PEE70 pKa = 3.96NMVKK74 pKa = 10.29RR75 pKa = 11.84MSEE78 pKa = 3.59AGKK81 pKa = 8.65RR82 pKa = 11.84TVEE85 pKa = 3.96ALVVKK90 pKa = 10.05YY91 pKa = 8.12QLKK94 pKa = 10.21SGNPGRR100 pKa = 11.84DD101 pKa = 3.51DD102 pKa = 3.36LTLARR107 pKa = 11.84VAAALAPWTCQAAFVVSEE125 pKa = 4.16FMPVTGRR132 pKa = 11.84AMDD135 pKa = 3.93DD136 pKa = 3.34HH137 pKa = 6.5AASYY141 pKa = 9.17PRR143 pKa = 11.84GMMHH147 pKa = 6.83TSFAGLIDD155 pKa = 3.56PTLPQSTYY163 pKa = 9.61SDD165 pKa = 3.49LVAAHH170 pKa = 6.89SLFLYY175 pKa = 10.08YY176 pKa = 10.47FSKK179 pKa = 9.82TINVGLRR186 pKa = 11.84GKK188 pKa = 8.6TKK190 pKa = 11.0AEE192 pKa = 3.97VEE194 pKa = 4.26ASFSQPMQAAINSGFISGSQRR215 pKa = 11.84RR216 pKa = 11.84QFLQNLGIIDD226 pKa = 4.18RR227 pKa = 11.84NSVAVPAVATAAAAYY242 pKa = 7.98RR243 pKa = 11.84ALSS246 pKa = 3.45
MM1 pKa = 7.2SADD4 pKa = 3.58YY5 pKa = 10.81QRR7 pKa = 11.84IAIEE11 pKa = 4.55FGSEE15 pKa = 3.99PIDD18 pKa = 3.8RR19 pKa = 11.84DD20 pKa = 5.19AITLWVNEE28 pKa = 3.91FAYY31 pKa = 10.56QGFDD35 pKa = 2.87ASNVLRR41 pKa = 11.84LLQQRR46 pKa = 11.84GGATWKK52 pKa = 10.48EE53 pKa = 4.07DD54 pKa = 3.36AKK56 pKa = 11.53KK57 pKa = 10.17MIILVLTRR65 pKa = 11.84GNKK68 pKa = 8.28PEE70 pKa = 3.96NMVKK74 pKa = 10.29RR75 pKa = 11.84MSEE78 pKa = 3.59AGKK81 pKa = 8.65RR82 pKa = 11.84TVEE85 pKa = 3.96ALVVKK90 pKa = 10.05YY91 pKa = 8.12QLKK94 pKa = 10.21SGNPGRR100 pKa = 11.84DD101 pKa = 3.51DD102 pKa = 3.36LTLARR107 pKa = 11.84VAAALAPWTCQAAFVVSEE125 pKa = 4.16FMPVTGRR132 pKa = 11.84AMDD135 pKa = 3.93DD136 pKa = 3.34HH137 pKa = 6.5AASYY141 pKa = 9.17PRR143 pKa = 11.84GMMHH147 pKa = 6.83TSFAGLIDD155 pKa = 3.56PTLPQSTYY163 pKa = 9.61SDD165 pKa = 3.49LVAAHH170 pKa = 6.89SLFLYY175 pKa = 10.08YY176 pKa = 10.47FSKK179 pKa = 9.82TINVGLRR186 pKa = 11.84GKK188 pKa = 8.6TKK190 pKa = 11.0AEE192 pKa = 3.97VEE194 pKa = 4.26ASFSQPMQAAINSGFISGSQRR215 pKa = 11.84RR216 pKa = 11.84QFLQNLGIIDD226 pKa = 4.18RR227 pKa = 11.84NSVAVPAVATAAAAYY242 pKa = 7.98RR243 pKa = 11.84ALSS246 pKa = 3.45
Molecular weight: 26.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4023 |
246 |
2090 |
1005.8 |
113.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.792 ± 1.443 | 2.784 ± 0.702 |
5.667 ± 0.486 | 6.513 ± 0.436 |
4.599 ± 0.317 | 5.841 ± 0.447 |
2.337 ± 0.381 | 6.985 ± 0.615 |
6.637 ± 0.537 | 8.949 ± 0.651 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.132 ± 0.68 | 3.853 ± 0.373 |
3.604 ± 0.278 | 3.082 ± 0.546 |
5.319 ± 0.377 | 10.241 ± 0.548 |
4.822 ± 0.205 | 5.866 ± 0.373 |
1.193 ± 0.146 | 2.784 ± 0.269 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
