
Camel associated porprismacovirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
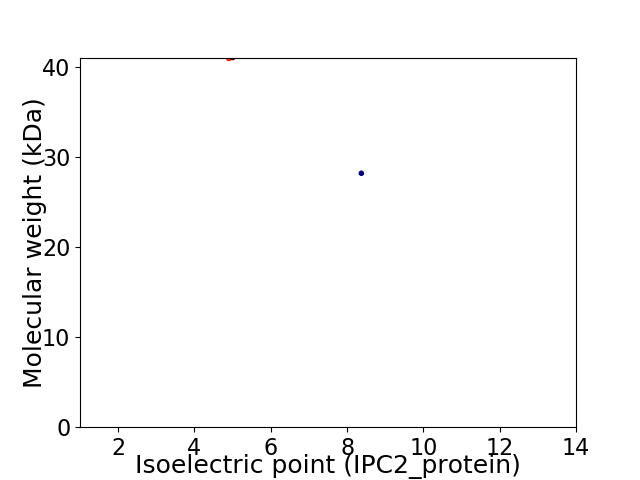
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1EL62|A0A0A1EL62_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 3 OX=2170107 GN=rep PE=4 SV=1
MM1 pKa = 7.45QITSVACIPLSEE13 pKa = 4.42GGKK16 pKa = 9.48IATNYY21 pKa = 7.24VTATYY26 pKa = 10.58QEE28 pKa = 4.52IYY30 pKa = 10.74DD31 pKa = 3.93LNTRR35 pKa = 11.84EE36 pKa = 4.45GKK38 pKa = 10.57LSIIGIHH45 pKa = 5.95TPTNIRR51 pKa = 11.84PIAQLQGFFAQYY63 pKa = 10.99KK64 pKa = 7.67KK65 pKa = 10.71FRR67 pKa = 11.84YY68 pKa = 9.73NGISKK73 pKa = 10.19IAIQPAAQLPADD85 pKa = 4.13PLQVSMEE92 pKa = 4.26AGQTLDD98 pKa = 4.89PRR100 pKa = 11.84DD101 pKa = 4.06LLNPILFHH109 pKa = 6.42GAHH112 pKa = 5.68GTSINAALNVAYY124 pKa = 10.12RR125 pKa = 11.84RR126 pKa = 11.84TGHH129 pKa = 5.8TFNTTSTNRR138 pKa = 11.84EE139 pKa = 3.98DD140 pKa = 3.06VAAIYY145 pKa = 10.24DD146 pKa = 3.71FSGDD150 pKa = 3.65EE151 pKa = 4.24TPVARR156 pKa = 11.84NLYY159 pKa = 9.73YY160 pKa = 10.51SALSDD165 pKa = 3.47PSFTKK170 pKa = 10.5FSPQQFIQLTDD181 pKa = 4.03LVPMVHH187 pKa = 6.69RR188 pKa = 11.84VNTSMYY194 pKa = 9.78FGPNQISEE202 pKa = 4.36MAGSNNGSAGTAINTEE218 pKa = 3.89IGRR221 pKa = 11.84ASQGTVWNGNQEE233 pKa = 4.19VTGPEE238 pKa = 4.1AIKK241 pKa = 11.0NEE243 pKa = 3.85WTLGFQQFFSNGMAPLDD260 pKa = 3.61WMPTRR265 pKa = 11.84TFNNMGDD272 pKa = 3.69VISTNEE278 pKa = 3.64NRR280 pKa = 11.84TNTEE284 pKa = 3.82MFTEE288 pKa = 4.75LPKK291 pKa = 10.88LYY293 pKa = 9.47MGVLIFPPSYY303 pKa = 9.86NQKK306 pKa = 10.35LYY308 pKa = 9.72FRR310 pKa = 11.84MVITHH315 pKa = 6.4SFSFKK320 pKa = 10.99DD321 pKa = 3.89FGTYY325 pKa = 8.44WSPEE329 pKa = 3.79MGDD332 pKa = 3.23WGLLEE337 pKa = 4.31YY338 pKa = 10.71FNEE341 pKa = 4.31FDD343 pKa = 5.0SSSTTQASSLNLTEE357 pKa = 4.83ADD359 pKa = 3.49AVLEE363 pKa = 4.11NDD365 pKa = 3.71GVMM368 pKa = 4.34
MM1 pKa = 7.45QITSVACIPLSEE13 pKa = 4.42GGKK16 pKa = 9.48IATNYY21 pKa = 7.24VTATYY26 pKa = 10.58QEE28 pKa = 4.52IYY30 pKa = 10.74DD31 pKa = 3.93LNTRR35 pKa = 11.84EE36 pKa = 4.45GKK38 pKa = 10.57LSIIGIHH45 pKa = 5.95TPTNIRR51 pKa = 11.84PIAQLQGFFAQYY63 pKa = 10.99KK64 pKa = 7.67KK65 pKa = 10.71FRR67 pKa = 11.84YY68 pKa = 9.73NGISKK73 pKa = 10.19IAIQPAAQLPADD85 pKa = 4.13PLQVSMEE92 pKa = 4.26AGQTLDD98 pKa = 4.89PRR100 pKa = 11.84DD101 pKa = 4.06LLNPILFHH109 pKa = 6.42GAHH112 pKa = 5.68GTSINAALNVAYY124 pKa = 10.12RR125 pKa = 11.84RR126 pKa = 11.84TGHH129 pKa = 5.8TFNTTSTNRR138 pKa = 11.84EE139 pKa = 3.98DD140 pKa = 3.06VAAIYY145 pKa = 10.24DD146 pKa = 3.71FSGDD150 pKa = 3.65EE151 pKa = 4.24TPVARR156 pKa = 11.84NLYY159 pKa = 9.73YY160 pKa = 10.51SALSDD165 pKa = 3.47PSFTKK170 pKa = 10.5FSPQQFIQLTDD181 pKa = 4.03LVPMVHH187 pKa = 6.69RR188 pKa = 11.84VNTSMYY194 pKa = 9.78FGPNQISEE202 pKa = 4.36MAGSNNGSAGTAINTEE218 pKa = 3.89IGRR221 pKa = 11.84ASQGTVWNGNQEE233 pKa = 4.19VTGPEE238 pKa = 4.1AIKK241 pKa = 11.0NEE243 pKa = 3.85WTLGFQQFFSNGMAPLDD260 pKa = 3.61WMPTRR265 pKa = 11.84TFNNMGDD272 pKa = 3.69VISTNEE278 pKa = 3.64NRR280 pKa = 11.84TNTEE284 pKa = 3.82MFTEE288 pKa = 4.75LPKK291 pKa = 10.88LYY293 pKa = 9.47MGVLIFPPSYY303 pKa = 9.86NQKK306 pKa = 10.35LYY308 pKa = 9.72FRR310 pKa = 11.84MVITHH315 pKa = 6.4SFSFKK320 pKa = 10.99DD321 pKa = 3.89FGTYY325 pKa = 8.44WSPEE329 pKa = 3.79MGDD332 pKa = 3.23WGLLEE337 pKa = 4.31YY338 pKa = 10.71FNEE341 pKa = 4.31FDD343 pKa = 5.0SSSTTQASSLNLTEE357 pKa = 4.83ADD359 pKa = 3.49AVLEE363 pKa = 4.11NDD365 pKa = 3.71GVMM368 pKa = 4.34
Molecular weight: 40.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1EL62|A0A0A1EL62_9VIRU Putative replicase protein OS=Camel associated porprismacovirus 3 OX=2170107 GN=rep PE=4 SV=1
MM1 pKa = 6.53VTVPTKK7 pKa = 10.15HH8 pKa = 5.09ACKK11 pKa = 9.91RR12 pKa = 11.84QVKK15 pKa = 9.19MILEE19 pKa = 4.17IFDD22 pKa = 3.42TKK24 pKa = 10.72RR25 pKa = 11.84YY26 pKa = 9.89IFAQEE31 pKa = 3.73KK32 pKa = 9.5GKK34 pKa = 10.55NGLDD38 pKa = 2.97HH39 pKa = 6.29WQLRR43 pKa = 11.84IQISGDD49 pKa = 3.05KK50 pKa = 10.49EE51 pKa = 4.23EE52 pKa = 4.32NFEE55 pKa = 4.09RR56 pKa = 11.84LQKK59 pKa = 10.52LFCNKK64 pKa = 9.31AHH66 pKa = 6.87IEE68 pKa = 4.16VANDD72 pKa = 2.45SWEE75 pKa = 4.04YY76 pKa = 9.95EE77 pKa = 4.04RR78 pKa = 11.84KK79 pKa = 9.67EE80 pKa = 3.98GKK82 pKa = 9.22FWSSEE87 pKa = 3.87DD88 pKa = 3.24TKK90 pKa = 11.07EE91 pKa = 3.62ILKK94 pKa = 10.16IRR96 pKa = 11.84FGKK99 pKa = 10.03LRR101 pKa = 11.84SEE103 pKa = 4.2QKK105 pKa = 10.57KK106 pKa = 9.68ILQILSSQGDD116 pKa = 3.59RR117 pKa = 11.84EE118 pKa = 3.93IDD120 pKa = 3.23VWLDD124 pKa = 3.08PTGCHH129 pKa = 5.5GKK131 pKa = 9.28SWLTIHH137 pKa = 6.61LWEE140 pKa = 4.39TGRR143 pKa = 11.84ALVVPRR149 pKa = 11.84SSTTAEE155 pKa = 3.92KK156 pKa = 10.77LSAFVCSSWRR166 pKa = 11.84GEE168 pKa = 4.18DD169 pKa = 3.31IIVIDD174 pKa = 4.07VPRR177 pKa = 11.84ATKK180 pKa = 8.66PTTSLLEE187 pKa = 4.11TMEE190 pKa = 4.13EE191 pKa = 4.16LKK193 pKa = 11.09DD194 pKa = 3.59GLVFDD199 pKa = 4.61HH200 pKa = 7.49RR201 pKa = 11.84YY202 pKa = 7.34TGKK205 pKa = 8.03TRR207 pKa = 11.84NVRR210 pKa = 11.84GTKK213 pKa = 10.57LMVFTNSPLPLNKK226 pKa = 10.16LSTDD230 pKa = 2.59RR231 pKa = 11.84WRR233 pKa = 11.84LHH235 pKa = 6.81GISADD240 pKa = 3.62GSLSS244 pKa = 3.34
MM1 pKa = 6.53VTVPTKK7 pKa = 10.15HH8 pKa = 5.09ACKK11 pKa = 9.91RR12 pKa = 11.84QVKK15 pKa = 9.19MILEE19 pKa = 4.17IFDD22 pKa = 3.42TKK24 pKa = 10.72RR25 pKa = 11.84YY26 pKa = 9.89IFAQEE31 pKa = 3.73KK32 pKa = 9.5GKK34 pKa = 10.55NGLDD38 pKa = 2.97HH39 pKa = 6.29WQLRR43 pKa = 11.84IQISGDD49 pKa = 3.05KK50 pKa = 10.49EE51 pKa = 4.23EE52 pKa = 4.32NFEE55 pKa = 4.09RR56 pKa = 11.84LQKK59 pKa = 10.52LFCNKK64 pKa = 9.31AHH66 pKa = 6.87IEE68 pKa = 4.16VANDD72 pKa = 2.45SWEE75 pKa = 4.04YY76 pKa = 9.95EE77 pKa = 4.04RR78 pKa = 11.84KK79 pKa = 9.67EE80 pKa = 3.98GKK82 pKa = 9.22FWSSEE87 pKa = 3.87DD88 pKa = 3.24TKK90 pKa = 11.07EE91 pKa = 3.62ILKK94 pKa = 10.16IRR96 pKa = 11.84FGKK99 pKa = 10.03LRR101 pKa = 11.84SEE103 pKa = 4.2QKK105 pKa = 10.57KK106 pKa = 9.68ILQILSSQGDD116 pKa = 3.59RR117 pKa = 11.84EE118 pKa = 3.93IDD120 pKa = 3.23VWLDD124 pKa = 3.08PTGCHH129 pKa = 5.5GKK131 pKa = 9.28SWLTIHH137 pKa = 6.61LWEE140 pKa = 4.39TGRR143 pKa = 11.84ALVVPRR149 pKa = 11.84SSTTAEE155 pKa = 3.92KK156 pKa = 10.77LSAFVCSSWRR166 pKa = 11.84GEE168 pKa = 4.18DD169 pKa = 3.31IIVIDD174 pKa = 4.07VPRR177 pKa = 11.84ATKK180 pKa = 8.66PTTSLLEE187 pKa = 4.11TMEE190 pKa = 4.13EE191 pKa = 4.16LKK193 pKa = 11.09DD194 pKa = 3.59GLVFDD199 pKa = 4.61HH200 pKa = 7.49RR201 pKa = 11.84YY202 pKa = 7.34TGKK205 pKa = 8.03TRR207 pKa = 11.84NVRR210 pKa = 11.84GTKK213 pKa = 10.57LMVFTNSPLPLNKK226 pKa = 10.16LSTDD230 pKa = 2.59RR231 pKa = 11.84WRR233 pKa = 11.84LHH235 pKa = 6.81GISADD240 pKa = 3.62GSLSS244 pKa = 3.34
Molecular weight: 28.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
612 |
244 |
368 |
306.0 |
34.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.882 ± 1.21 | 0.817 ± 0.453 |
4.902 ± 0.461 | 6.209 ± 0.87 |
5.065 ± 0.759 | 6.863 ± 0.394 |
2.124 ± 0.411 | 6.373 ± 0.102 |
5.392 ± 2.224 | 8.17 ± 0.919 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.628 | 5.719 ± 1.571 |
4.412 ± 0.851 | 4.412 ± 0.625 |
4.902 ± 1.139 | 7.68 ± 0.059 |
8.497 ± 0.617 | 4.739 ± 0.325 |
2.124 ± 0.637 | 2.941 ± 0.944 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
