
Apis mellifera associated microvirus 24
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
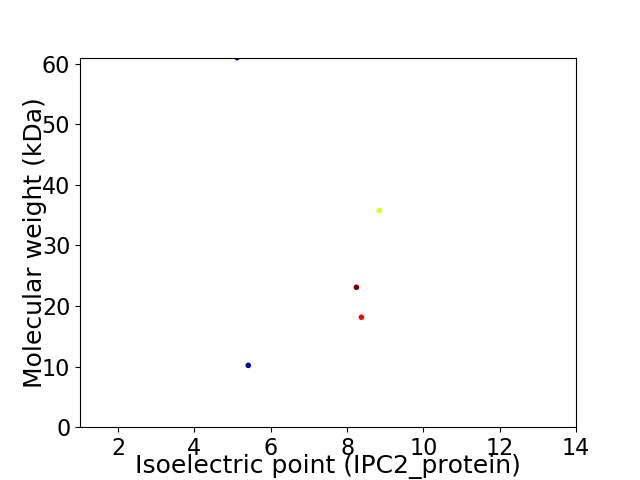
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q8U5Q7|A0A3Q8U5Q7_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 24 OX=2494752 PE=3 SV=1
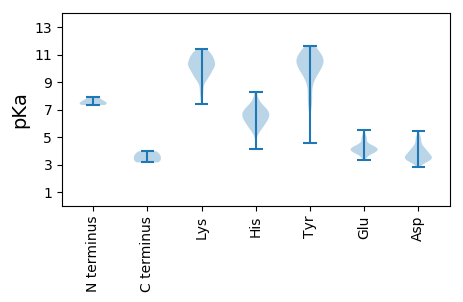
MM1 pKa = 7.37NLGSRR6 pKa = 11.84YY7 pKa = 8.46SQHH10 pKa = 6.61SFAQIPDD17 pKa = 3.26VNMARR22 pKa = 11.84SKK24 pKa = 10.55FNRR27 pKa = 11.84SFAVKK32 pKa = 9.34DD33 pKa = 3.39TFNFDD38 pKa = 3.49DD39 pKa = 4.76LVPFFIDD46 pKa = 4.33EE47 pKa = 5.08IIPGDD52 pKa = 4.04TINLNVKK59 pKa = 9.77SFSRR63 pKa = 11.84LAPQVVPLMDD73 pKa = 4.32NMYY76 pKa = 9.42MDD78 pKa = 3.79YY79 pKa = 11.13FFFFVPNRR87 pKa = 11.84LVWNNWEE94 pKa = 4.4KK95 pKa = 11.01FCGAQDD101 pKa = 4.2DD102 pKa = 4.97PGDD105 pKa = 3.82STDD108 pKa = 3.91YY109 pKa = 10.3IVPRR113 pKa = 11.84IALAEE118 pKa = 4.03TGANVVGSIYY128 pKa = 10.71DD129 pKa = 3.37KK130 pKa = 11.16FGIPTDD136 pKa = 3.87VPDD139 pKa = 3.6VTINALPFRR148 pKa = 11.84AYY150 pKa = 11.2NLIFNSWFRR159 pKa = 11.84DD160 pKa = 3.4QNLQDD165 pKa = 4.02SLPVPKK171 pKa = 9.86TDD173 pKa = 5.11GPDD176 pKa = 3.14NPADD180 pKa = 3.78YY181 pKa = 10.63IIVQRR186 pKa = 11.84GKK188 pKa = 9.15RR189 pKa = 11.84HH190 pKa = 7.0DD191 pKa = 4.22YY192 pKa = 8.15FTSALPWPQKK202 pKa = 10.85GEE204 pKa = 3.86AVTIPTGGLAPVVGTITGPSSLAVLPYY231 pKa = 10.02RR232 pKa = 11.84ANTGAFPDD240 pKa = 3.55ANIRR244 pKa = 11.84SGVASTTGTAIFASGTSLPNNTDD267 pKa = 2.81INPYY271 pKa = 9.75ISAGGLTMSHH281 pKa = 5.91TLEE284 pKa = 4.92ADD286 pKa = 3.43LTEE289 pKa = 4.42STAITINQFRR299 pKa = 11.84LAMMTQSFLEE309 pKa = 3.97LDD311 pKa = 3.34ARR313 pKa = 11.84GGTRR317 pKa = 11.84YY318 pKa = 10.04VEE320 pKa = 4.23ILRR323 pKa = 11.84AHH325 pKa = 6.15YY326 pKa = 10.55NVVSPDD332 pKa = 3.2FRR334 pKa = 11.84LQRR337 pKa = 11.84PEE339 pKa = 4.0LLSVGSSTISQHH351 pKa = 6.31VVAQNSATTEE361 pKa = 3.88TSPQGNLAAFSTAYY375 pKa = 9.37TGSSRR380 pKa = 11.84IGFSKK385 pKa = 10.71SFVEE389 pKa = 3.84HH390 pKa = 6.77GYY392 pKa = 11.01VIGLVAARR400 pKa = 11.84ADD402 pKa = 3.48LTYY405 pKa = 10.67QQGLNRR411 pKa = 11.84MWSRR415 pKa = 11.84TTRR418 pKa = 11.84QDD420 pKa = 3.34FFWPKK425 pKa = 9.43FQEE428 pKa = 4.26LGEE431 pKa = 3.86QAIFNRR437 pKa = 11.84EE438 pKa = 3.33IFYY441 pKa = 11.13SGVAEE446 pKa = 5.28DD447 pKa = 4.51DD448 pKa = 4.02DD449 pKa = 5.47AVFGYY454 pKa = 7.42QEE456 pKa = 4.17RR457 pKa = 11.84YY458 pKa = 9.58AEE460 pKa = 4.06YY461 pKa = 9.72RR462 pKa = 11.84YY463 pKa = 10.26KK464 pKa = 10.54PSEE467 pKa = 3.37IRR469 pKa = 11.84GQFRR473 pKa = 11.84STYY476 pKa = 11.11AEE478 pKa = 4.14TLDD481 pKa = 3.5VWHH484 pKa = 6.78LAEE487 pKa = 5.01EE488 pKa = 5.01FSSTPALNGEE498 pKa = 4.81FIKK501 pKa = 10.85SSTPIEE507 pKa = 4.22RR508 pKa = 11.84SLVVPNPTYY517 pKa = 10.34PHH519 pKa = 7.13LLFDD523 pKa = 3.63CWFDD527 pKa = 3.73YY528 pKa = 10.77KK529 pKa = 10.44HH530 pKa = 6.35ARR532 pKa = 11.84PMTTYY537 pKa = 10.53GVPATLGRR545 pKa = 11.84FF546 pKa = 3.54
MM1 pKa = 7.37NLGSRR6 pKa = 11.84YY7 pKa = 8.46SQHH10 pKa = 6.61SFAQIPDD17 pKa = 3.26VNMARR22 pKa = 11.84SKK24 pKa = 10.55FNRR27 pKa = 11.84SFAVKK32 pKa = 9.34DD33 pKa = 3.39TFNFDD38 pKa = 3.49DD39 pKa = 4.76LVPFFIDD46 pKa = 4.33EE47 pKa = 5.08IIPGDD52 pKa = 4.04TINLNVKK59 pKa = 9.77SFSRR63 pKa = 11.84LAPQVVPLMDD73 pKa = 4.32NMYY76 pKa = 9.42MDD78 pKa = 3.79YY79 pKa = 11.13FFFFVPNRR87 pKa = 11.84LVWNNWEE94 pKa = 4.4KK95 pKa = 11.01FCGAQDD101 pKa = 4.2DD102 pKa = 4.97PGDD105 pKa = 3.82STDD108 pKa = 3.91YY109 pKa = 10.3IVPRR113 pKa = 11.84IALAEE118 pKa = 4.03TGANVVGSIYY128 pKa = 10.71DD129 pKa = 3.37KK130 pKa = 11.16FGIPTDD136 pKa = 3.87VPDD139 pKa = 3.6VTINALPFRR148 pKa = 11.84AYY150 pKa = 11.2NLIFNSWFRR159 pKa = 11.84DD160 pKa = 3.4QNLQDD165 pKa = 4.02SLPVPKK171 pKa = 9.86TDD173 pKa = 5.11GPDD176 pKa = 3.14NPADD180 pKa = 3.78YY181 pKa = 10.63IIVQRR186 pKa = 11.84GKK188 pKa = 9.15RR189 pKa = 11.84HH190 pKa = 7.0DD191 pKa = 4.22YY192 pKa = 8.15FTSALPWPQKK202 pKa = 10.85GEE204 pKa = 3.86AVTIPTGGLAPVVGTITGPSSLAVLPYY231 pKa = 10.02RR232 pKa = 11.84ANTGAFPDD240 pKa = 3.55ANIRR244 pKa = 11.84SGVASTTGTAIFASGTSLPNNTDD267 pKa = 2.81INPYY271 pKa = 9.75ISAGGLTMSHH281 pKa = 5.91TLEE284 pKa = 4.92ADD286 pKa = 3.43LTEE289 pKa = 4.42STAITINQFRR299 pKa = 11.84LAMMTQSFLEE309 pKa = 3.97LDD311 pKa = 3.34ARR313 pKa = 11.84GGTRR317 pKa = 11.84YY318 pKa = 10.04VEE320 pKa = 4.23ILRR323 pKa = 11.84AHH325 pKa = 6.15YY326 pKa = 10.55NVVSPDD332 pKa = 3.2FRR334 pKa = 11.84LQRR337 pKa = 11.84PEE339 pKa = 4.0LLSVGSSTISQHH351 pKa = 6.31VVAQNSATTEE361 pKa = 3.88TSPQGNLAAFSTAYY375 pKa = 9.37TGSSRR380 pKa = 11.84IGFSKK385 pKa = 10.71SFVEE389 pKa = 3.84HH390 pKa = 6.77GYY392 pKa = 11.01VIGLVAARR400 pKa = 11.84ADD402 pKa = 3.48LTYY405 pKa = 10.67QQGLNRR411 pKa = 11.84MWSRR415 pKa = 11.84TTRR418 pKa = 11.84QDD420 pKa = 3.34FFWPKK425 pKa = 9.43FQEE428 pKa = 4.26LGEE431 pKa = 3.86QAIFNRR437 pKa = 11.84EE438 pKa = 3.33IFYY441 pKa = 11.13SGVAEE446 pKa = 5.28DD447 pKa = 4.51DD448 pKa = 4.02DD449 pKa = 5.47AVFGYY454 pKa = 7.42QEE456 pKa = 4.17RR457 pKa = 11.84YY458 pKa = 9.58AEE460 pKa = 4.06YY461 pKa = 9.72RR462 pKa = 11.84YY463 pKa = 10.26KK464 pKa = 10.54PSEE467 pKa = 3.37IRR469 pKa = 11.84GQFRR473 pKa = 11.84STYY476 pKa = 11.11AEE478 pKa = 4.14TLDD481 pKa = 3.5VWHH484 pKa = 6.78LAEE487 pKa = 5.01EE488 pKa = 5.01FSSTPALNGEE498 pKa = 4.81FIKK501 pKa = 10.85SSTPIEE507 pKa = 4.22RR508 pKa = 11.84SLVVPNPTYY517 pKa = 10.34PHH519 pKa = 7.13LLFDD523 pKa = 3.63CWFDD527 pKa = 3.73YY528 pKa = 10.77KK529 pKa = 10.44HH530 pKa = 6.35ARR532 pKa = 11.84PMTTYY537 pKa = 10.53GVPATLGRR545 pKa = 11.84FF546 pKa = 3.54
Molecular weight: 60.99 kDa
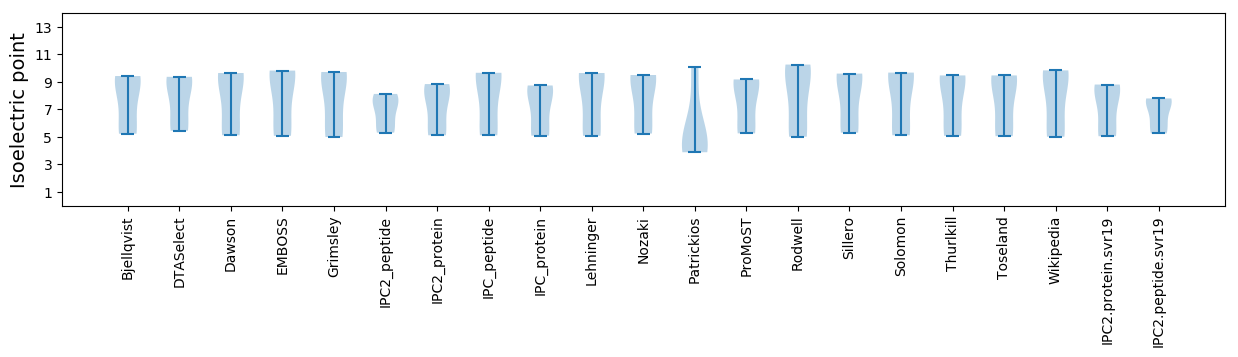
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U5N3|A0A3Q8U5N3_9VIRU Nonstructural protein OS=Apis mellifera associated microvirus 24 OX=2494752 PE=4 SV=1
PP1 pKa = 7.33AGNRR5 pKa = 11.84IYY7 pKa = 10.7SPKK10 pKa = 10.13NLPPGLVPDD19 pKa = 4.04VFSCRR24 pKa = 11.84KK25 pKa = 9.39CLPCRR30 pKa = 11.84LNNAKK35 pKa = 9.99EE36 pKa = 3.67KK37 pKa = 10.59AIRR40 pKa = 11.84CWHH43 pKa = 5.99EE44 pKa = 4.37SKK46 pKa = 9.16MHH48 pKa = 7.39DD49 pKa = 4.03DD50 pKa = 4.93NIFLTLTYY58 pKa = 10.32SDD60 pKa = 4.12EE61 pKa = 4.31HH62 pKa = 8.25LEE64 pKa = 4.21SPRR67 pKa = 11.84LIYY70 pKa = 9.35SHH72 pKa = 5.37WQEE75 pKa = 4.91FIHH78 pKa = 7.04KK79 pKa = 9.48LRR81 pKa = 11.84DD82 pKa = 3.46KK83 pKa = 10.73IRR85 pKa = 11.84YY86 pKa = 8.73NSLGEE91 pKa = 4.07QKK93 pKa = 10.14ISTMVTGEE101 pKa = 3.92YY102 pKa = 10.85GEE104 pKa = 4.26QTKK107 pKa = 10.24RR108 pKa = 11.84PHH110 pKa = 4.12WHH112 pKa = 6.62ALIFNYY118 pKa = 10.17RR119 pKa = 11.84PGDD122 pKa = 3.5EE123 pKa = 4.01RR124 pKa = 11.84PYY126 pKa = 10.01RR127 pKa = 11.84TTEE130 pKa = 3.94RR131 pKa = 11.84GDD133 pKa = 3.13KK134 pKa = 10.23VYY136 pKa = 10.6RR137 pKa = 11.84SEE139 pKa = 4.91TIQTLWNKK147 pKa = 10.37GYY149 pKa = 10.16IEE151 pKa = 4.56YY152 pKa = 10.99GDD154 pKa = 3.73VTIDD158 pKa = 3.24SANYY162 pKa = 7.17VARR165 pKa = 11.84YY166 pKa = 7.72AAKK169 pKa = 10.32KK170 pKa = 9.26LVHH173 pKa = 6.9GIDD176 pKa = 3.45QAHH179 pKa = 7.67DD180 pKa = 3.31YY181 pKa = 10.95HH182 pKa = 6.95PIHH185 pKa = 6.33RR186 pKa = 11.84TSSKK190 pKa = 10.29HH191 pKa = 6.32AIGKK195 pKa = 9.2RR196 pKa = 11.84WIEE199 pKa = 3.88KK200 pKa = 9.82YY201 pKa = 10.03YY202 pKa = 10.79KK203 pKa = 8.71QTFEE207 pKa = 4.1HH208 pKa = 6.98GYY210 pKa = 9.85VLLPNFQKK218 pKa = 11.12AKK220 pKa = 9.46IPRR223 pKa = 11.84YY224 pKa = 7.54YY225 pKa = 10.61CEE227 pKa = 4.12WVEE230 pKa = 4.09KK231 pKa = 9.73HH232 pKa = 6.6HH233 pKa = 5.73PAIWEE238 pKa = 4.17RR239 pKa = 11.84YY240 pKa = 4.57VTQIRR245 pKa = 11.84PRR247 pKa = 11.84LQEE250 pKa = 3.44QAEE253 pKa = 3.99AAEE256 pKa = 4.47RR257 pKa = 11.84AEE259 pKa = 4.2EE260 pKa = 4.48MIFLTNLINKK270 pKa = 9.39SCGAPHH276 pKa = 6.65PMKK279 pKa = 10.41RR280 pKa = 11.84SQVKK284 pKa = 7.98EE285 pKa = 4.23TILKK289 pKa = 10.61AKK291 pKa = 10.15FKK293 pKa = 10.57QLQEE297 pKa = 4.03RR298 pKa = 11.84LKK300 pKa = 11.11LL301 pKa = 3.72
PP1 pKa = 7.33AGNRR5 pKa = 11.84IYY7 pKa = 10.7SPKK10 pKa = 10.13NLPPGLVPDD19 pKa = 4.04VFSCRR24 pKa = 11.84KK25 pKa = 9.39CLPCRR30 pKa = 11.84LNNAKK35 pKa = 9.99EE36 pKa = 3.67KK37 pKa = 10.59AIRR40 pKa = 11.84CWHH43 pKa = 5.99EE44 pKa = 4.37SKK46 pKa = 9.16MHH48 pKa = 7.39DD49 pKa = 4.03DD50 pKa = 4.93NIFLTLTYY58 pKa = 10.32SDD60 pKa = 4.12EE61 pKa = 4.31HH62 pKa = 8.25LEE64 pKa = 4.21SPRR67 pKa = 11.84LIYY70 pKa = 9.35SHH72 pKa = 5.37WQEE75 pKa = 4.91FIHH78 pKa = 7.04KK79 pKa = 9.48LRR81 pKa = 11.84DD82 pKa = 3.46KK83 pKa = 10.73IRR85 pKa = 11.84YY86 pKa = 8.73NSLGEE91 pKa = 4.07QKK93 pKa = 10.14ISTMVTGEE101 pKa = 3.92YY102 pKa = 10.85GEE104 pKa = 4.26QTKK107 pKa = 10.24RR108 pKa = 11.84PHH110 pKa = 4.12WHH112 pKa = 6.62ALIFNYY118 pKa = 10.17RR119 pKa = 11.84PGDD122 pKa = 3.5EE123 pKa = 4.01RR124 pKa = 11.84PYY126 pKa = 10.01RR127 pKa = 11.84TTEE130 pKa = 3.94RR131 pKa = 11.84GDD133 pKa = 3.13KK134 pKa = 10.23VYY136 pKa = 10.6RR137 pKa = 11.84SEE139 pKa = 4.91TIQTLWNKK147 pKa = 10.37GYY149 pKa = 10.16IEE151 pKa = 4.56YY152 pKa = 10.99GDD154 pKa = 3.73VTIDD158 pKa = 3.24SANYY162 pKa = 7.17VARR165 pKa = 11.84YY166 pKa = 7.72AAKK169 pKa = 10.32KK170 pKa = 9.26LVHH173 pKa = 6.9GIDD176 pKa = 3.45QAHH179 pKa = 7.67DD180 pKa = 3.31YY181 pKa = 10.95HH182 pKa = 6.95PIHH185 pKa = 6.33RR186 pKa = 11.84TSSKK190 pKa = 10.29HH191 pKa = 6.32AIGKK195 pKa = 9.2RR196 pKa = 11.84WIEE199 pKa = 3.88KK200 pKa = 9.82YY201 pKa = 10.03YY202 pKa = 10.79KK203 pKa = 8.71QTFEE207 pKa = 4.1HH208 pKa = 6.98GYY210 pKa = 9.85VLLPNFQKK218 pKa = 11.12AKK220 pKa = 9.46IPRR223 pKa = 11.84YY224 pKa = 7.54YY225 pKa = 10.61CEE227 pKa = 4.12WVEE230 pKa = 4.09KK231 pKa = 9.73HH232 pKa = 6.6HH233 pKa = 5.73PAIWEE238 pKa = 4.17RR239 pKa = 11.84YY240 pKa = 4.57VTQIRR245 pKa = 11.84PRR247 pKa = 11.84LQEE250 pKa = 3.44QAEE253 pKa = 3.99AAEE256 pKa = 4.47RR257 pKa = 11.84AEE259 pKa = 4.2EE260 pKa = 4.48MIFLTNLINKK270 pKa = 9.39SCGAPHH276 pKa = 6.65PMKK279 pKa = 10.41RR280 pKa = 11.84SQVKK284 pKa = 7.98EE285 pKa = 4.23TILKK289 pKa = 10.61AKK291 pKa = 10.15FKK293 pKa = 10.57QLQEE297 pKa = 4.03RR298 pKa = 11.84LKK300 pKa = 11.11LL301 pKa = 3.72
Molecular weight: 35.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1302 |
87 |
546 |
260.4 |
29.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.22 ± 0.86 | 0.845 ± 0.32 |
5.991 ± 0.904 | 5.76 ± 0.832 |
4.916 ± 1.013 | 5.223 ± 0.666 |
2.381 ± 0.799 | 5.069 ± 0.772 |
5.914 ± 1.651 | 7.143 ± 0.241 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.843 ± 0.281 | 5.223 ± 0.512 |
6.375 ± 0.612 | 4.916 ± 0.791 |
6.298 ± 0.487 | 7.45 ± 1.106 |
6.682 ± 0.623 | 5.146 ± 0.642 |
1.459 ± 0.269 | 4.147 ± 0.857 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
