
Pyrococcus abyssi virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified archaeal dsDNA viruses
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
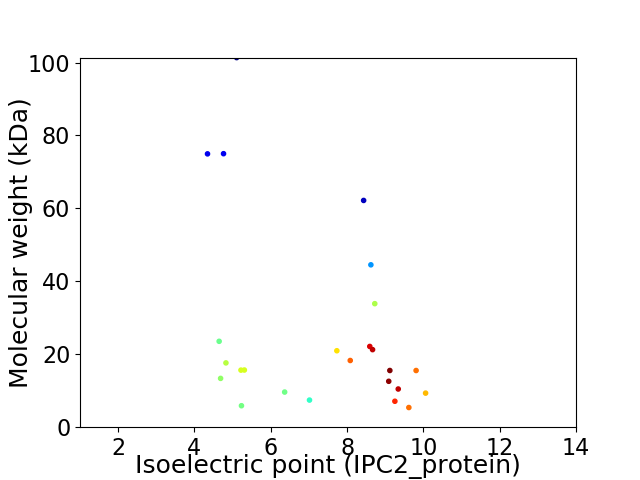
Virtual 2D-PAGE plot for 25 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A5Y2D8|A5Y2D8_9VIRU Membrane protein OS=Pyrococcus abyssi virus 1 OX=425386 GN=ORF138 PE=4 SV=1
MM1 pKa = 7.23TRR3 pKa = 11.84RR4 pKa = 11.84LSALLIIILAGLLAPQVKK22 pKa = 9.87AATTSYY28 pKa = 11.15YY29 pKa = 10.66GVQFDD34 pKa = 4.51GVDD37 pKa = 3.72DD38 pKa = 3.93YY39 pKa = 12.35VSFNSVTLDD48 pKa = 3.47ANASTVIIFFRR59 pKa = 11.84VNAFSDD65 pKa = 3.42SAYY68 pKa = 10.63GEE70 pKa = 4.09RR71 pKa = 11.84ANIISKK77 pKa = 10.78DD78 pKa = 3.33SGKK81 pKa = 9.65YY82 pKa = 8.8NSYY85 pKa = 10.71IGFTSSGIYY94 pKa = 10.5AEE96 pKa = 4.9GNTNGDD102 pKa = 2.71YY103 pKa = 10.02WAIYY107 pKa = 9.12PYY109 pKa = 10.16TLNLGKK115 pKa = 9.02TYY117 pKa = 10.29MLALVADD124 pKa = 4.59SEE126 pKa = 4.75QVTTYY131 pKa = 11.51LDD133 pKa = 3.52TTKK136 pKa = 10.94LGTRR140 pKa = 11.84PVLSNMTVNALGGAYY155 pKa = 9.97AGSDD159 pKa = 3.09NFFDD163 pKa = 3.82GKK165 pKa = 10.52IYY167 pKa = 10.8LVLIYY172 pKa = 10.64NRR174 pKa = 11.84ALSEE178 pKa = 4.29SEE180 pKa = 3.48IQAIYY185 pKa = 9.95EE186 pKa = 4.42DD187 pKa = 4.56PSNPPTSGLVLWYY200 pKa = 10.39APDD203 pKa = 3.75SVDD206 pKa = 3.16PSTNTWHH213 pKa = 7.32DD214 pKa = 3.26KK215 pKa = 10.81SGNGHH220 pKa = 7.06DD221 pKa = 3.74GSIYY225 pKa = 9.69GATYY229 pKa = 10.49VPLRR233 pKa = 11.84PISEE237 pKa = 4.58SEE239 pKa = 3.77FSDD242 pKa = 3.74AYY244 pKa = 10.5GVAFGSGDD252 pKa = 3.61YY253 pKa = 10.64VVASNVVTGKK263 pKa = 9.99QLTAIVFASVDD274 pKa = 3.24KK275 pKa = 10.7FYY277 pKa = 11.09TPDD280 pKa = 4.01DD281 pKa = 4.39PNKK284 pKa = 10.22GLQPLVSKK292 pKa = 11.05GSDD295 pKa = 2.85RR296 pKa = 11.84FKK298 pKa = 11.11LYY300 pKa = 10.92VGGNYY305 pKa = 10.09ARR307 pKa = 11.84FILVDD312 pKa = 3.3NSLNVYY318 pKa = 10.07RR319 pKa = 11.84LIAGAVEE326 pKa = 4.23NEE328 pKa = 3.84VFMLAGVVDD337 pKa = 3.56NTIAKK342 pKa = 9.75IYY344 pKa = 10.72LNGEE348 pKa = 4.04LKK350 pKa = 9.56EE351 pKa = 4.41TEE353 pKa = 4.26TLSDD357 pKa = 3.91VISDD361 pKa = 4.1YY362 pKa = 11.72VDD364 pKa = 3.07TADD367 pKa = 3.36IFLGKK372 pKa = 10.03RR373 pKa = 11.84SPGDD377 pKa = 3.51YY378 pKa = 10.02QQLDD382 pKa = 3.53GKK384 pKa = 10.38IYY386 pKa = 10.81LVLIYY391 pKa = 10.66NRR393 pKa = 11.84ALSDD397 pKa = 3.58SEE399 pKa = 3.67IRR401 pKa = 11.84QIYY404 pKa = 7.9EE405 pKa = 3.94NPSNPPLDD413 pKa = 5.2GLILWYY419 pKa = 10.08DD420 pKa = 4.06PYY422 pKa = 11.45SYY424 pKa = 11.19DD425 pKa = 3.45PSTGKK430 pKa = 9.09WLNRR434 pKa = 11.84APIFPTIPLVEE445 pKa = 4.29EE446 pKa = 4.31LDD448 pKa = 3.86GVNYY452 pKa = 9.76GAEE455 pKa = 4.18AVRR458 pKa = 11.84VSIPAVKK465 pKa = 10.22AYY467 pKa = 10.01DD468 pKa = 3.45ATDD471 pKa = 3.24NSEE474 pKa = 3.56ISVYY478 pKa = 11.05NLTIKK483 pKa = 10.76SIDD486 pKa = 3.66GEE488 pKa = 4.67TFIPWFMPLPYY499 pKa = 10.48NQTLTLNVTAPGYY512 pKa = 9.85KK513 pKa = 9.61SAIISVQAPIDD524 pKa = 3.52NLVFYY529 pKa = 9.07LYY531 pKa = 9.35PSQVITSWEE540 pKa = 3.91SAQININLNNITADD554 pKa = 3.34EE555 pKa = 4.37FLPDD559 pKa = 3.9ASQDD563 pKa = 2.89IDD565 pKa = 3.69KK566 pKa = 10.71FSSLFTRR573 pKa = 11.84WLGGEE578 pKa = 4.21YY579 pKa = 10.08FKK581 pKa = 11.25HH582 pKa = 6.23LLSLDD587 pKa = 3.82LEE589 pKa = 4.7GMLKK593 pKa = 10.64DD594 pKa = 4.42FFNNPPIPGLGILVALMIWAASVLMTWLYY623 pKa = 10.8FEE625 pKa = 4.66EE626 pKa = 4.86PEE628 pKa = 4.2PVFVEE633 pKa = 4.17GNIVYY638 pKa = 10.16FGLMQFIDD646 pKa = 4.02VSPGILAVPLGVFILFLGYY665 pKa = 9.75KK666 pKa = 9.32QLYY669 pKa = 8.97KK670 pKa = 10.72ALKK673 pKa = 8.52VWAKK677 pKa = 10.18RR678 pKa = 3.54
MM1 pKa = 7.23TRR3 pKa = 11.84RR4 pKa = 11.84LSALLIIILAGLLAPQVKK22 pKa = 9.87AATTSYY28 pKa = 11.15YY29 pKa = 10.66GVQFDD34 pKa = 4.51GVDD37 pKa = 3.72DD38 pKa = 3.93YY39 pKa = 12.35VSFNSVTLDD48 pKa = 3.47ANASTVIIFFRR59 pKa = 11.84VNAFSDD65 pKa = 3.42SAYY68 pKa = 10.63GEE70 pKa = 4.09RR71 pKa = 11.84ANIISKK77 pKa = 10.78DD78 pKa = 3.33SGKK81 pKa = 9.65YY82 pKa = 8.8NSYY85 pKa = 10.71IGFTSSGIYY94 pKa = 10.5AEE96 pKa = 4.9GNTNGDD102 pKa = 2.71YY103 pKa = 10.02WAIYY107 pKa = 9.12PYY109 pKa = 10.16TLNLGKK115 pKa = 9.02TYY117 pKa = 10.29MLALVADD124 pKa = 4.59SEE126 pKa = 4.75QVTTYY131 pKa = 11.51LDD133 pKa = 3.52TTKK136 pKa = 10.94LGTRR140 pKa = 11.84PVLSNMTVNALGGAYY155 pKa = 9.97AGSDD159 pKa = 3.09NFFDD163 pKa = 3.82GKK165 pKa = 10.52IYY167 pKa = 10.8LVLIYY172 pKa = 10.64NRR174 pKa = 11.84ALSEE178 pKa = 4.29SEE180 pKa = 3.48IQAIYY185 pKa = 9.95EE186 pKa = 4.42DD187 pKa = 4.56PSNPPTSGLVLWYY200 pKa = 10.39APDD203 pKa = 3.75SVDD206 pKa = 3.16PSTNTWHH213 pKa = 7.32DD214 pKa = 3.26KK215 pKa = 10.81SGNGHH220 pKa = 7.06DD221 pKa = 3.74GSIYY225 pKa = 9.69GATYY229 pKa = 10.49VPLRR233 pKa = 11.84PISEE237 pKa = 4.58SEE239 pKa = 3.77FSDD242 pKa = 3.74AYY244 pKa = 10.5GVAFGSGDD252 pKa = 3.61YY253 pKa = 10.64VVASNVVTGKK263 pKa = 9.99QLTAIVFASVDD274 pKa = 3.24KK275 pKa = 10.7FYY277 pKa = 11.09TPDD280 pKa = 4.01DD281 pKa = 4.39PNKK284 pKa = 10.22GLQPLVSKK292 pKa = 11.05GSDD295 pKa = 2.85RR296 pKa = 11.84FKK298 pKa = 11.11LYY300 pKa = 10.92VGGNYY305 pKa = 10.09ARR307 pKa = 11.84FILVDD312 pKa = 3.3NSLNVYY318 pKa = 10.07RR319 pKa = 11.84LIAGAVEE326 pKa = 4.23NEE328 pKa = 3.84VFMLAGVVDD337 pKa = 3.56NTIAKK342 pKa = 9.75IYY344 pKa = 10.72LNGEE348 pKa = 4.04LKK350 pKa = 9.56EE351 pKa = 4.41TEE353 pKa = 4.26TLSDD357 pKa = 3.91VISDD361 pKa = 4.1YY362 pKa = 11.72VDD364 pKa = 3.07TADD367 pKa = 3.36IFLGKK372 pKa = 10.03RR373 pKa = 11.84SPGDD377 pKa = 3.51YY378 pKa = 10.02QQLDD382 pKa = 3.53GKK384 pKa = 10.38IYY386 pKa = 10.81LVLIYY391 pKa = 10.66NRR393 pKa = 11.84ALSDD397 pKa = 3.58SEE399 pKa = 3.67IRR401 pKa = 11.84QIYY404 pKa = 7.9EE405 pKa = 3.94NPSNPPLDD413 pKa = 5.2GLILWYY419 pKa = 10.08DD420 pKa = 4.06PYY422 pKa = 11.45SYY424 pKa = 11.19DD425 pKa = 3.45PSTGKK430 pKa = 9.09WLNRR434 pKa = 11.84APIFPTIPLVEE445 pKa = 4.29EE446 pKa = 4.31LDD448 pKa = 3.86GVNYY452 pKa = 9.76GAEE455 pKa = 4.18AVRR458 pKa = 11.84VSIPAVKK465 pKa = 10.22AYY467 pKa = 10.01DD468 pKa = 3.45ATDD471 pKa = 3.24NSEE474 pKa = 3.56ISVYY478 pKa = 11.05NLTIKK483 pKa = 10.76SIDD486 pKa = 3.66GEE488 pKa = 4.67TFIPWFMPLPYY499 pKa = 10.48NQTLTLNVTAPGYY512 pKa = 9.85KK513 pKa = 9.61SAIISVQAPIDD524 pKa = 3.52NLVFYY529 pKa = 9.07LYY531 pKa = 9.35PSQVITSWEE540 pKa = 3.91SAQININLNNITADD554 pKa = 3.34EE555 pKa = 4.37FLPDD559 pKa = 3.9ASQDD563 pKa = 2.89IDD565 pKa = 3.69KK566 pKa = 10.71FSSLFTRR573 pKa = 11.84WLGGEE578 pKa = 4.21YY579 pKa = 10.08FKK581 pKa = 11.25HH582 pKa = 6.23LLSLDD587 pKa = 3.82LEE589 pKa = 4.7GMLKK593 pKa = 10.64DD594 pKa = 4.42FFNNPPIPGLGILVALMIWAASVLMTWLYY623 pKa = 10.8FEE625 pKa = 4.66EE626 pKa = 4.86PEE628 pKa = 4.2PVFVEE633 pKa = 4.17GNIVYY638 pKa = 10.16FGLMQFIDD646 pKa = 4.02VSPGILAVPLGVFILFLGYY665 pKa = 9.75KK666 pKa = 9.32QLYY669 pKa = 8.97KK670 pKa = 10.72ALKK673 pKa = 8.52VWAKK677 pKa = 10.18RR678 pKa = 3.54
Molecular weight: 74.95 kDa
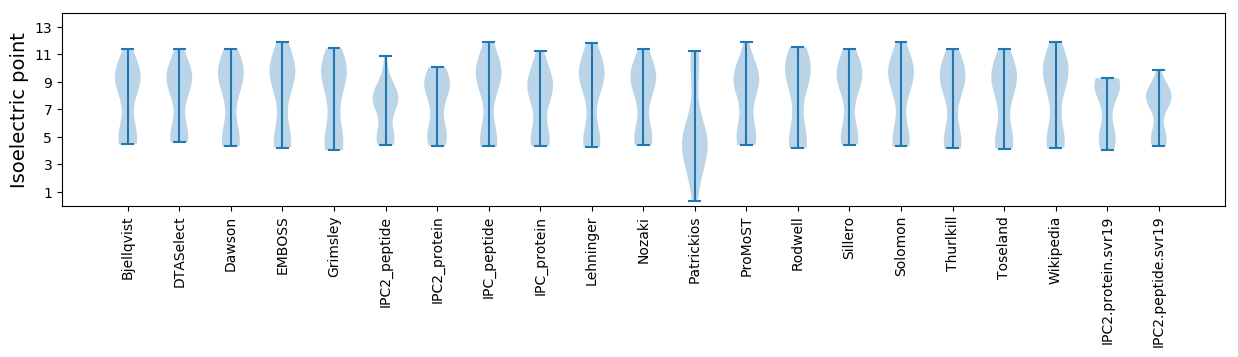
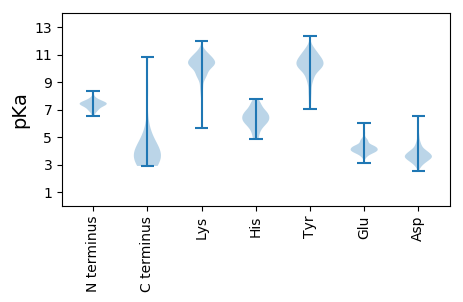
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A5Y2D0|A5Y2D0_9VIRU Membrane protein OS=Pyrococcus abyssi virus 1 OX=425386 GN=ORF121 PE=4 SV=1
MM1 pKa = 8.38DD2 pKa = 5.53IPPLLCGGQGPIVGPTRR19 pKa = 11.84KK20 pKa = 9.59QYY22 pKa = 8.35ATKK25 pKa = 10.03IFKK28 pKa = 10.38HH29 pKa = 6.02FSSMGNNYY37 pKa = 9.6HH38 pKa = 6.62LRR40 pKa = 11.84EE41 pKa = 3.9KK42 pKa = 10.53KK43 pKa = 9.8RR44 pKa = 11.84YY45 pKa = 8.12KK46 pKa = 10.37GKK48 pKa = 10.47SRR50 pKa = 11.84EE51 pKa = 4.22TQNTPSSSRR60 pKa = 11.84KK61 pKa = 8.55PNQSAYY67 pKa = 7.7ITKK70 pKa = 10.32HH71 pKa = 5.92KK72 pKa = 8.48NTKK75 pKa = 10.09RR76 pKa = 11.84KK77 pKa = 9.55ILGVNN82 pKa = 3.57
MM1 pKa = 8.38DD2 pKa = 5.53IPPLLCGGQGPIVGPTRR19 pKa = 11.84KK20 pKa = 9.59QYY22 pKa = 8.35ATKK25 pKa = 10.03IFKK28 pKa = 10.38HH29 pKa = 6.02FSSMGNNYY37 pKa = 9.6HH38 pKa = 6.62LRR40 pKa = 11.84EE41 pKa = 3.9KK42 pKa = 10.53KK43 pKa = 9.8RR44 pKa = 11.84YY45 pKa = 8.12KK46 pKa = 10.37GKK48 pKa = 10.47SRR50 pKa = 11.84EE51 pKa = 4.22TQNTPSSSRR60 pKa = 11.84KK61 pKa = 8.55PNQSAYY67 pKa = 7.7ITKK70 pKa = 10.32HH71 pKa = 5.92KK72 pKa = 8.48NTKK75 pKa = 10.09RR76 pKa = 11.84KK77 pKa = 9.55ILGVNN82 pKa = 3.57
Molecular weight: 9.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5787 |
52 |
898 |
231.5 |
26.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.757 ± 0.364 | 0.156 ± 0.064 |
5.754 ± 0.713 | 6.722 ± 0.781 |
4.942 ± 0.368 | 6.29 ± 0.5 |
1.002 ± 0.178 | 7.223 ± 0.299 |
7.189 ± 0.789 | 11.007 ± 0.648 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.357 | 3.819 ± 0.304 |
3.819 ± 0.398 | 2.989 ± 0.23 |
5.149 ± 0.589 | 5.772 ± 0.496 |
5.132 ± 0.602 | 7.482 ± 0.417 |
1.728 ± 0.203 | 4.579 ± 0.506 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
