
Aerococcus urinaehominis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Aerococcaceae; Aerococcus
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
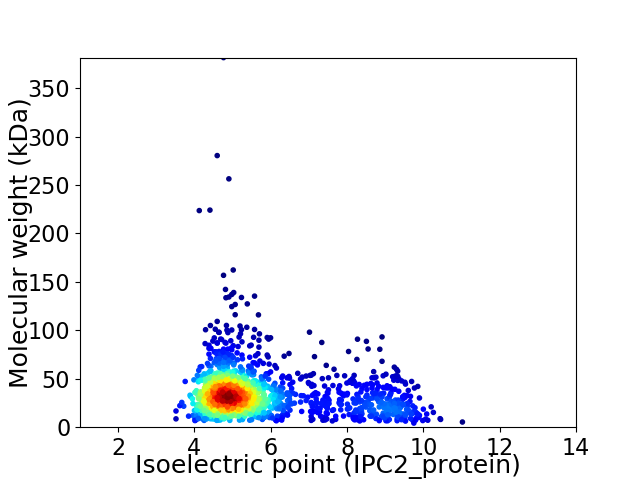
Virtual 2D-PAGE plot for 1595 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0X8FLI7|A0A0X8FLI7_9LACT ATP-dependent helicase/nuclease subunit A OS=Aerococcus urinaehominis OX=128944 GN=addA PE=3 SV=1
MM1 pKa = 7.12SFSVKK6 pKa = 10.67SMLGTSLATTAALVAIAPSVVADD29 pKa = 4.87DD30 pKa = 4.58YY31 pKa = 10.86TIQWGDD37 pKa = 3.42TLTDD41 pKa = 4.04LAQKK45 pKa = 10.83HH46 pKa = 5.41NVTLDD51 pKa = 3.39DD52 pKa = 4.32LKK54 pKa = 11.08AANGITDD61 pKa = 3.59SGDD64 pKa = 3.79LIIAGHH70 pKa = 5.5TLVIPEE76 pKa = 4.74LPQVVGQSQATDD88 pKa = 3.5GQNVYY93 pKa = 8.46TVQAGDD99 pKa = 3.83TLNKK103 pKa = 9.76IAAQFGTTADD113 pKa = 3.49SLRR116 pKa = 11.84ALNNIQGDD124 pKa = 4.47LIFIGQQLVVAGQVAEE140 pKa = 4.25AATVAPVVEE149 pKa = 4.8EE150 pKa = 3.9PAVAEE155 pKa = 4.18EE156 pKa = 4.54TTVEE160 pKa = 3.95AEE162 pKa = 4.1PAVVEE167 pKa = 4.06TDD169 pKa = 3.35KK170 pKa = 11.57LDD172 pKa = 3.86EE173 pKa = 5.05AEE175 pKa = 4.25LAQAVAYY182 pKa = 9.62QAEE185 pKa = 4.61EE186 pKa = 3.88VAPIAEE192 pKa = 4.34EE193 pKa = 4.04LVAPASEE200 pKa = 3.98AVGEE204 pKa = 4.22QEE206 pKa = 4.82VAATSSNEE214 pKa = 3.99VEE216 pKa = 4.31STEE219 pKa = 3.92ATVTEE224 pKa = 4.53EE225 pKa = 4.1VAPKK229 pKa = 10.72AEE231 pKa = 4.52AKK233 pKa = 10.56EE234 pKa = 4.06EE235 pKa = 4.45TPVATAEE242 pKa = 4.1TAAAAPASAEE252 pKa = 3.91ATAVDD257 pKa = 4.32AEE259 pKa = 4.42VAPAVQAAPAPAQEE273 pKa = 4.33TVTVTEE279 pKa = 4.83AEE281 pKa = 4.07TAPIEE286 pKa = 4.07EE287 pKa = 4.85AAPVAQPVAYY297 pKa = 7.78EE298 pKa = 4.15APAVEE303 pKa = 4.59APAPVKK309 pKa = 10.67EE310 pKa = 4.08EE311 pKa = 4.29APAEE315 pKa = 4.04TAPVEE320 pKa = 4.29EE321 pKa = 4.57EE322 pKa = 4.42VVAEE326 pKa = 4.29PVQEE330 pKa = 4.07AAPAAEE336 pKa = 4.28TQHH339 pKa = 6.33NGSSVLEE346 pKa = 4.26VADD349 pKa = 3.73QYY351 pKa = 12.04VGTPYY356 pKa = 10.63VWGGRR361 pKa = 11.84QPGGFDD367 pKa = 3.08CSGFVQYY374 pKa = 10.51VYY376 pKa = 10.85RR377 pKa = 11.84EE378 pKa = 4.07AEE380 pKa = 4.01GRR382 pKa = 11.84EE383 pKa = 4.06VGSWTGEE390 pKa = 3.7QQYY393 pKa = 10.8AGPQIAVEE401 pKa = 4.33SAQQGDD407 pKa = 3.9LLFWGDD413 pKa = 3.63YY414 pKa = 9.95GNPYY418 pKa = 9.59HH419 pKa = 6.62VAISTGDD426 pKa = 3.01NGYY429 pKa = 9.11IHH431 pKa = 7.01SPQPGQTVGYY441 pKa = 8.05GTVSEE446 pKa = 4.24YY447 pKa = 9.95WYY449 pKa = 9.93PSFAVQMM456 pKa = 4.47
MM1 pKa = 7.12SFSVKK6 pKa = 10.67SMLGTSLATTAALVAIAPSVVADD29 pKa = 4.87DD30 pKa = 4.58YY31 pKa = 10.86TIQWGDD37 pKa = 3.42TLTDD41 pKa = 4.04LAQKK45 pKa = 10.83HH46 pKa = 5.41NVTLDD51 pKa = 3.39DD52 pKa = 4.32LKK54 pKa = 11.08AANGITDD61 pKa = 3.59SGDD64 pKa = 3.79LIIAGHH70 pKa = 5.5TLVIPEE76 pKa = 4.74LPQVVGQSQATDD88 pKa = 3.5GQNVYY93 pKa = 8.46TVQAGDD99 pKa = 3.83TLNKK103 pKa = 9.76IAAQFGTTADD113 pKa = 3.49SLRR116 pKa = 11.84ALNNIQGDD124 pKa = 4.47LIFIGQQLVVAGQVAEE140 pKa = 4.25AATVAPVVEE149 pKa = 4.8EE150 pKa = 3.9PAVAEE155 pKa = 4.18EE156 pKa = 4.54TTVEE160 pKa = 3.95AEE162 pKa = 4.1PAVVEE167 pKa = 4.06TDD169 pKa = 3.35KK170 pKa = 11.57LDD172 pKa = 3.86EE173 pKa = 5.05AEE175 pKa = 4.25LAQAVAYY182 pKa = 9.62QAEE185 pKa = 4.61EE186 pKa = 3.88VAPIAEE192 pKa = 4.34EE193 pKa = 4.04LVAPASEE200 pKa = 3.98AVGEE204 pKa = 4.22QEE206 pKa = 4.82VAATSSNEE214 pKa = 3.99VEE216 pKa = 4.31STEE219 pKa = 3.92ATVTEE224 pKa = 4.53EE225 pKa = 4.1VAPKK229 pKa = 10.72AEE231 pKa = 4.52AKK233 pKa = 10.56EE234 pKa = 4.06EE235 pKa = 4.45TPVATAEE242 pKa = 4.1TAAAAPASAEE252 pKa = 3.91ATAVDD257 pKa = 4.32AEE259 pKa = 4.42VAPAVQAAPAPAQEE273 pKa = 4.33TVTVTEE279 pKa = 4.83AEE281 pKa = 4.07TAPIEE286 pKa = 4.07EE287 pKa = 4.85AAPVAQPVAYY297 pKa = 7.78EE298 pKa = 4.15APAVEE303 pKa = 4.59APAPVKK309 pKa = 10.67EE310 pKa = 4.08EE311 pKa = 4.29APAEE315 pKa = 4.04TAPVEE320 pKa = 4.29EE321 pKa = 4.57EE322 pKa = 4.42VVAEE326 pKa = 4.29PVQEE330 pKa = 4.07AAPAAEE336 pKa = 4.28TQHH339 pKa = 6.33NGSSVLEE346 pKa = 4.26VADD349 pKa = 3.73QYY351 pKa = 12.04VGTPYY356 pKa = 10.63VWGGRR361 pKa = 11.84QPGGFDD367 pKa = 3.08CSGFVQYY374 pKa = 10.51VYY376 pKa = 10.85RR377 pKa = 11.84EE378 pKa = 4.07AEE380 pKa = 4.01GRR382 pKa = 11.84EE383 pKa = 4.06VGSWTGEE390 pKa = 3.7QQYY393 pKa = 10.8AGPQIAVEE401 pKa = 4.33SAQQGDD407 pKa = 3.9LLFWGDD413 pKa = 3.63YY414 pKa = 9.95GNPYY418 pKa = 9.59HH419 pKa = 6.62VAISTGDD426 pKa = 3.01NGYY429 pKa = 9.11IHH431 pKa = 7.01SPQPGQTVGYY441 pKa = 8.05GTVSEE446 pKa = 4.24YY447 pKa = 9.95WYY449 pKa = 9.93PSFAVQMM456 pKa = 4.47
Molecular weight: 47.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0X8FMZ0|A0A0X8FMZ0_9LACT O-antigen polymerase OS=Aerococcus urinaehominis OX=128944 GN=AWM75_05845 PE=4 SV=1
MM1 pKa = 7.46LRR3 pKa = 11.84ALDD6 pKa = 3.37KK7 pKa = 11.11HH8 pKa = 6.08GLVKK12 pKa = 10.64GLMMGTGRR20 pKa = 11.84ILRR23 pKa = 11.84CHH25 pKa = 6.51PFARR29 pKa = 11.84GGFDD33 pKa = 3.28PVPDD37 pKa = 3.63HH38 pKa = 6.08FTLRR42 pKa = 11.84RR43 pKa = 11.84NRR45 pKa = 11.84SNDD48 pKa = 3.01LPEE51 pKa = 5.48AEE53 pKa = 4.39KK54 pKa = 10.5QALIADD60 pKa = 3.78HH61 pKa = 6.58HH62 pKa = 6.56RR63 pKa = 11.84RR64 pKa = 11.84HH65 pKa = 5.77HH66 pKa = 5.89HH67 pKa = 5.5
MM1 pKa = 7.46LRR3 pKa = 11.84ALDD6 pKa = 3.37KK7 pKa = 11.11HH8 pKa = 6.08GLVKK12 pKa = 10.64GLMMGTGRR20 pKa = 11.84ILRR23 pKa = 11.84CHH25 pKa = 6.51PFARR29 pKa = 11.84GGFDD33 pKa = 3.28PVPDD37 pKa = 3.63HH38 pKa = 6.08FTLRR42 pKa = 11.84RR43 pKa = 11.84NRR45 pKa = 11.84SNDD48 pKa = 3.01LPEE51 pKa = 5.48AEE53 pKa = 4.39KK54 pKa = 10.5QALIADD60 pKa = 3.78HH61 pKa = 6.58HH62 pKa = 6.56RR63 pKa = 11.84RR64 pKa = 11.84HH65 pKa = 5.77HH66 pKa = 5.89HH67 pKa = 5.5
Molecular weight: 7.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
516534 |
37 |
3575 |
323.8 |
36.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.881 ± 0.09 | 0.594 ± 0.017 |
6.687 ± 0.058 | 5.898 ± 0.063 |
3.937 ± 0.047 | 6.859 ± 0.066 |
2.002 ± 0.027 | 6.959 ± 0.059 |
5.234 ± 0.067 | 9.986 ± 0.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.495 ± 0.04 | 4.373 ± 0.041 |
3.694 ± 0.052 | 5.747 ± 0.075 |
4.204 ± 0.044 | 5.725 ± 0.042 |
5.131 ± 0.083 | 6.879 ± 0.05 |
0.868 ± 0.023 | 3.849 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
