
Flavobacterium dankookense
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
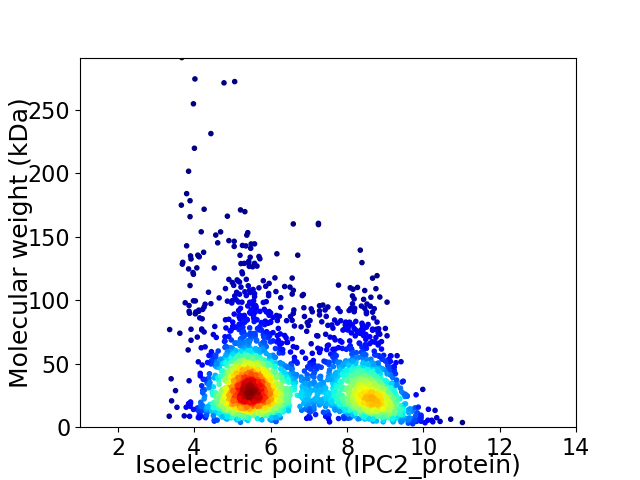
Virtual 2D-PAGE plot for 2803 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6QAK8|A0A4R6QAK8_9FLAO N-acylneuraminate cytidylyltransferase OS=Flavobacterium dankookense OX=706186 GN=BC748_1463 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.4KK3 pKa = 8.43ITNLCKK9 pKa = 10.45LLILFGITFTYY20 pKa = 10.39AQTARR25 pKa = 11.84VQVIHH30 pKa = 6.62NSPDD34 pKa = 3.11LAAQEE39 pKa = 3.77VDD41 pKa = 3.57VYY43 pKa = 11.36INGTIALDD51 pKa = 3.35NFAFRR56 pKa = 11.84TATPFIDD63 pKa = 4.73LPAHH67 pKa = 6.04APITIDD73 pKa = 3.32VAPASSTSVLEE84 pKa = 4.24TLSTTTTTLTAGEE97 pKa = 4.36TYY99 pKa = 10.13IIVANGIVSPTGYY112 pKa = 10.21SVAPNFALSVYY123 pKa = 10.24SGGRR127 pKa = 11.84EE128 pKa = 4.0GTPVLTEE135 pKa = 3.73TDD137 pKa = 3.46VLVAHH142 pKa = 7.18GSPDD146 pKa = 3.52APTVDD151 pKa = 3.58VVEE154 pKa = 4.47TGVGAGTIVNNLSYY168 pKa = 11.32GSFSNYY174 pKa = 10.31LEE176 pKa = 4.38LTTANYY182 pKa = 10.07ILDD185 pKa = 3.72VRR187 pKa = 11.84DD188 pKa = 3.3EE189 pKa = 4.45TGTVTVARR197 pKa = 11.84YY198 pKa = 7.82SAPLSDD204 pKa = 6.17LFLQSDD210 pKa = 5.24AITVVASGFLNPDD223 pKa = 3.45NNSDD227 pKa = 3.74GPAFGLWVAKK237 pKa = 9.25ATGGALIPLPLYY249 pKa = 10.86NPTAKK254 pKa = 10.45VQVIHH259 pKa = 6.55NSPDD263 pKa = 3.47AIAAEE268 pKa = 3.92VDD270 pKa = 3.77VYY272 pKa = 11.48INGDD276 pKa = 3.55LALDD280 pKa = 3.47NFAFRR285 pKa = 11.84TATPFIDD292 pKa = 4.95LPAEE296 pKa = 3.93TSISIDD302 pKa = 3.16IAPSTSSSSSEE313 pKa = 4.01SIYY316 pKa = 11.36NLTTSLAANEE326 pKa = 4.32TYY328 pKa = 10.21IIVANGIVSPTGYY341 pKa = 10.21SVAPNFALNVFSQGRR356 pKa = 11.84EE357 pKa = 3.74FADD360 pKa = 3.89DD361 pKa = 3.96SNLVDD366 pKa = 3.35VLVNHH371 pKa = 7.29GSPDD375 pKa = 3.55APTVDD380 pKa = 3.58VVEE383 pKa = 4.4TAVPAGTVVDD393 pKa = 4.5NISYY397 pKa = 9.08PQFAGYY403 pKa = 10.58LSLPEE408 pKa = 4.98ADD410 pKa = 3.94FVLDD414 pKa = 3.71VRR416 pKa = 11.84DD417 pKa = 3.65EE418 pKa = 4.39TGATTVASYY427 pKa = 9.23QAPFNTFNYY436 pKa = 9.12AGSAITVLASGFLNPAANSDD456 pKa = 3.81GPAFGLWVAFSNGGPLFEE474 pKa = 5.37LPLVEE479 pKa = 4.53VPQTARR485 pKa = 11.84LQVIHH490 pKa = 6.66NSPDD494 pKa = 3.47AIAAEE499 pKa = 3.88VDD501 pKa = 3.86VYY503 pKa = 12.02VNGTLTLDD511 pKa = 3.2NFAFRR516 pKa = 11.84TATPFIDD523 pKa = 3.95VPAGVALSIDD533 pKa = 3.74VAPGNSTSATEE544 pKa = 3.99SVYY547 pKa = 11.39NLTTTLADD555 pKa = 3.56GEE557 pKa = 4.72TYY559 pKa = 10.24IAIANGIVSPTGYY572 pKa = 10.21SVAPNFALSVFAQGRR587 pKa = 11.84EE588 pKa = 4.18VASDD592 pKa = 3.57PAEE595 pKa = 3.92TDD597 pKa = 3.26VLVNHH602 pKa = 7.3GSPDD606 pKa = 3.55APTVDD611 pKa = 3.58VVEE614 pKa = 4.47TGVGAGTIVNDD625 pKa = 3.39ISYY628 pKa = 8.91PQFAGYY634 pKa = 10.64LEE636 pKa = 4.77LPNLDD641 pKa = 3.35FVLDD645 pKa = 3.71VRR647 pKa = 11.84DD648 pKa = 3.77ATGTTTVARR657 pKa = 11.84YY658 pKa = 7.84SAPLQTLNLDD668 pKa = 3.88GAALTVVASGFLNPAANSDD687 pKa = 3.81GPAFGLWVATAAGGNLIQLPPFVEE711 pKa = 4.4PTARR715 pKa = 11.84LQVIHH720 pKa = 6.66NSPDD724 pKa = 3.47AIAAEE729 pKa = 3.88VDD731 pKa = 3.86VYY733 pKa = 12.02VNGTLTLDD741 pKa = 3.2NFAFRR746 pKa = 11.84TATPFIDD753 pKa = 3.95VPAGVALSIDD763 pKa = 3.74VAPGNSTSATEE774 pKa = 4.12SIYY777 pKa = 11.37NLTTTLADD785 pKa = 3.56GEE787 pKa = 4.72TYY789 pKa = 10.24IAIANGIVSPTGYY802 pKa = 10.21SVAPNFALSVFAQGRR817 pKa = 11.84EE818 pKa = 4.04AASDD822 pKa = 3.65PAEE825 pKa = 3.97TDD827 pKa = 3.26VLVNHH832 pKa = 7.3GSPDD836 pKa = 3.55APTVDD841 pKa = 3.58VVEE844 pKa = 4.47TGVGAGTIVNDD855 pKa = 3.39ISYY858 pKa = 8.91PQFAGYY864 pKa = 10.64LEE866 pKa = 4.77LPNLDD871 pKa = 3.35FVLDD875 pKa = 3.71VRR877 pKa = 11.84DD878 pKa = 3.77ATGTTTVASYY888 pKa = 9.76LAPLQTLNLDD898 pKa = 3.73GAAITVIASGFLNPAANSDD917 pKa = 3.81GPAFGLWVATAAGGNLIQLPPFVEE941 pKa = 4.4PTARR945 pKa = 11.84LQVIHH950 pKa = 6.66NSPDD954 pKa = 3.47AIAAEE959 pKa = 3.88VDD961 pKa = 3.86VYY963 pKa = 12.02VNGTLTLDD971 pKa = 3.2NFAFRR976 pKa = 11.84TATPFIDD983 pKa = 3.95VPAGVALSIDD993 pKa = 3.74VAPGNSTSATEE1004 pKa = 3.99SVYY1007 pKa = 11.39NLTTTLADD1015 pKa = 3.56GEE1017 pKa = 4.72TYY1019 pKa = 10.24IAIANGIVSPTGYY1032 pKa = 10.21SVAPNFALSVFAQGRR1047 pKa = 11.84EE1048 pKa = 4.04AASDD1052 pKa = 3.65PAEE1055 pKa = 3.97TDD1057 pKa = 3.26VLVNHH1062 pKa = 7.3GSPDD1066 pKa = 3.55APTVDD1071 pKa = 3.58VVEE1074 pKa = 4.47TGVGAGTIVNDD1085 pKa = 3.39ISYY1088 pKa = 8.91PQFAGYY1094 pKa = 10.64LEE1096 pKa = 4.77LPNLDD1101 pKa = 3.35FVLDD1105 pKa = 3.71VRR1107 pKa = 11.84DD1108 pKa = 3.77ATGTTTVASYY1118 pKa = 9.76LAPLQTLNLDD1128 pKa = 3.73GAAITVIASGFLNPAVNSDD1147 pKa = 3.45GPAFGLWVATAAGGNLIPLPTAPLSVNEE1175 pKa = 4.55NDD1177 pKa = 3.88LEE1179 pKa = 4.68SIRR1182 pKa = 11.84IYY1184 pKa = 10.38PNPANSIINIDD1195 pKa = 3.53IPFSYY1200 pKa = 10.84DD1201 pKa = 2.96NSKK1204 pKa = 10.13VRR1206 pKa = 11.84LFDD1209 pKa = 3.53VNGRR1213 pKa = 11.84SIKK1216 pKa = 10.41EE1217 pKa = 3.9VTSVNSIDD1225 pKa = 3.66VTGLGSGIYY1234 pKa = 9.03MLSLEE1239 pKa = 4.66IDD1241 pKa = 3.19NTVINKK1247 pKa = 10.12KK1248 pKa = 10.01IVITNN1253 pKa = 3.86
MM1 pKa = 7.57KK2 pKa = 10.4KK3 pKa = 8.43ITNLCKK9 pKa = 10.45LLILFGITFTYY20 pKa = 10.39AQTARR25 pKa = 11.84VQVIHH30 pKa = 6.62NSPDD34 pKa = 3.11LAAQEE39 pKa = 3.77VDD41 pKa = 3.57VYY43 pKa = 11.36INGTIALDD51 pKa = 3.35NFAFRR56 pKa = 11.84TATPFIDD63 pKa = 4.73LPAHH67 pKa = 6.04APITIDD73 pKa = 3.32VAPASSTSVLEE84 pKa = 4.24TLSTTTTTLTAGEE97 pKa = 4.36TYY99 pKa = 10.13IIVANGIVSPTGYY112 pKa = 10.21SVAPNFALSVYY123 pKa = 10.24SGGRR127 pKa = 11.84EE128 pKa = 4.0GTPVLTEE135 pKa = 3.73TDD137 pKa = 3.46VLVAHH142 pKa = 7.18GSPDD146 pKa = 3.52APTVDD151 pKa = 3.58VVEE154 pKa = 4.47TGVGAGTIVNNLSYY168 pKa = 11.32GSFSNYY174 pKa = 10.31LEE176 pKa = 4.38LTTANYY182 pKa = 10.07ILDD185 pKa = 3.72VRR187 pKa = 11.84DD188 pKa = 3.3EE189 pKa = 4.45TGTVTVARR197 pKa = 11.84YY198 pKa = 7.82SAPLSDD204 pKa = 6.17LFLQSDD210 pKa = 5.24AITVVASGFLNPDD223 pKa = 3.45NNSDD227 pKa = 3.74GPAFGLWVAKK237 pKa = 9.25ATGGALIPLPLYY249 pKa = 10.86NPTAKK254 pKa = 10.45VQVIHH259 pKa = 6.55NSPDD263 pKa = 3.47AIAAEE268 pKa = 3.92VDD270 pKa = 3.77VYY272 pKa = 11.48INGDD276 pKa = 3.55LALDD280 pKa = 3.47NFAFRR285 pKa = 11.84TATPFIDD292 pKa = 4.95LPAEE296 pKa = 3.93TSISIDD302 pKa = 3.16IAPSTSSSSSEE313 pKa = 4.01SIYY316 pKa = 11.36NLTTSLAANEE326 pKa = 4.32TYY328 pKa = 10.21IIVANGIVSPTGYY341 pKa = 10.21SVAPNFALNVFSQGRR356 pKa = 11.84EE357 pKa = 3.74FADD360 pKa = 3.89DD361 pKa = 3.96SNLVDD366 pKa = 3.35VLVNHH371 pKa = 7.29GSPDD375 pKa = 3.55APTVDD380 pKa = 3.58VVEE383 pKa = 4.4TAVPAGTVVDD393 pKa = 4.5NISYY397 pKa = 9.08PQFAGYY403 pKa = 10.58LSLPEE408 pKa = 4.98ADD410 pKa = 3.94FVLDD414 pKa = 3.71VRR416 pKa = 11.84DD417 pKa = 3.65EE418 pKa = 4.39TGATTVASYY427 pKa = 9.23QAPFNTFNYY436 pKa = 9.12AGSAITVLASGFLNPAANSDD456 pKa = 3.81GPAFGLWVAFSNGGPLFEE474 pKa = 5.37LPLVEE479 pKa = 4.53VPQTARR485 pKa = 11.84LQVIHH490 pKa = 6.66NSPDD494 pKa = 3.47AIAAEE499 pKa = 3.88VDD501 pKa = 3.86VYY503 pKa = 12.02VNGTLTLDD511 pKa = 3.2NFAFRR516 pKa = 11.84TATPFIDD523 pKa = 3.95VPAGVALSIDD533 pKa = 3.74VAPGNSTSATEE544 pKa = 3.99SVYY547 pKa = 11.39NLTTTLADD555 pKa = 3.56GEE557 pKa = 4.72TYY559 pKa = 10.24IAIANGIVSPTGYY572 pKa = 10.21SVAPNFALSVFAQGRR587 pKa = 11.84EE588 pKa = 4.18VASDD592 pKa = 3.57PAEE595 pKa = 3.92TDD597 pKa = 3.26VLVNHH602 pKa = 7.3GSPDD606 pKa = 3.55APTVDD611 pKa = 3.58VVEE614 pKa = 4.47TGVGAGTIVNDD625 pKa = 3.39ISYY628 pKa = 8.91PQFAGYY634 pKa = 10.64LEE636 pKa = 4.77LPNLDD641 pKa = 3.35FVLDD645 pKa = 3.71VRR647 pKa = 11.84DD648 pKa = 3.77ATGTTTVARR657 pKa = 11.84YY658 pKa = 7.84SAPLQTLNLDD668 pKa = 3.88GAALTVVASGFLNPAANSDD687 pKa = 3.81GPAFGLWVATAAGGNLIQLPPFVEE711 pKa = 4.4PTARR715 pKa = 11.84LQVIHH720 pKa = 6.66NSPDD724 pKa = 3.47AIAAEE729 pKa = 3.88VDD731 pKa = 3.86VYY733 pKa = 12.02VNGTLTLDD741 pKa = 3.2NFAFRR746 pKa = 11.84TATPFIDD753 pKa = 3.95VPAGVALSIDD763 pKa = 3.74VAPGNSTSATEE774 pKa = 4.12SIYY777 pKa = 11.37NLTTTLADD785 pKa = 3.56GEE787 pKa = 4.72TYY789 pKa = 10.24IAIANGIVSPTGYY802 pKa = 10.21SVAPNFALSVFAQGRR817 pKa = 11.84EE818 pKa = 4.04AASDD822 pKa = 3.65PAEE825 pKa = 3.97TDD827 pKa = 3.26VLVNHH832 pKa = 7.3GSPDD836 pKa = 3.55APTVDD841 pKa = 3.58VVEE844 pKa = 4.47TGVGAGTIVNDD855 pKa = 3.39ISYY858 pKa = 8.91PQFAGYY864 pKa = 10.64LEE866 pKa = 4.77LPNLDD871 pKa = 3.35FVLDD875 pKa = 3.71VRR877 pKa = 11.84DD878 pKa = 3.77ATGTTTVASYY888 pKa = 9.76LAPLQTLNLDD898 pKa = 3.73GAAITVIASGFLNPAANSDD917 pKa = 3.81GPAFGLWVATAAGGNLIQLPPFVEE941 pKa = 4.4PTARR945 pKa = 11.84LQVIHH950 pKa = 6.66NSPDD954 pKa = 3.47AIAAEE959 pKa = 3.88VDD961 pKa = 3.86VYY963 pKa = 12.02VNGTLTLDD971 pKa = 3.2NFAFRR976 pKa = 11.84TATPFIDD983 pKa = 3.95VPAGVALSIDD993 pKa = 3.74VAPGNSTSATEE1004 pKa = 3.99SVYY1007 pKa = 11.39NLTTTLADD1015 pKa = 3.56GEE1017 pKa = 4.72TYY1019 pKa = 10.24IAIANGIVSPTGYY1032 pKa = 10.21SVAPNFALSVFAQGRR1047 pKa = 11.84EE1048 pKa = 4.04AASDD1052 pKa = 3.65PAEE1055 pKa = 3.97TDD1057 pKa = 3.26VLVNHH1062 pKa = 7.3GSPDD1066 pKa = 3.55APTVDD1071 pKa = 3.58VVEE1074 pKa = 4.47TGVGAGTIVNDD1085 pKa = 3.39ISYY1088 pKa = 8.91PQFAGYY1094 pKa = 10.64LEE1096 pKa = 4.77LPNLDD1101 pKa = 3.35FVLDD1105 pKa = 3.71VRR1107 pKa = 11.84DD1108 pKa = 3.77ATGTTTVASYY1118 pKa = 9.76LAPLQTLNLDD1128 pKa = 3.73GAAITVIASGFLNPAVNSDD1147 pKa = 3.45GPAFGLWVATAAGGNLIPLPTAPLSVNEE1175 pKa = 4.55NDD1177 pKa = 3.88LEE1179 pKa = 4.68SIRR1182 pKa = 11.84IYY1184 pKa = 10.38PNPANSIINIDD1195 pKa = 3.53IPFSYY1200 pKa = 10.84DD1201 pKa = 2.96NSKK1204 pKa = 10.13VRR1206 pKa = 11.84LFDD1209 pKa = 3.53VNGRR1213 pKa = 11.84SIKK1216 pKa = 10.41EE1217 pKa = 3.9VTSVNSIDD1225 pKa = 3.66VTGLGSGIYY1234 pKa = 9.03MLSLEE1239 pKa = 4.66IDD1241 pKa = 3.19NTVINKK1247 pKa = 10.12KK1248 pKa = 10.01IVITNN1253 pKa = 3.86
Molecular weight: 129.84 kDa
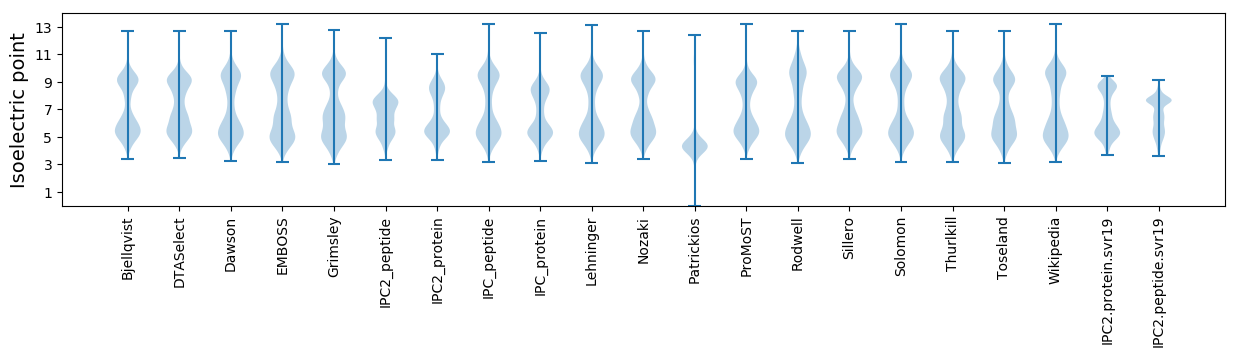
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6QCJ7|A0A4R6QCJ7_9FLAO Uncharacterized protein OS=Flavobacterium dankookense OX=706186 GN=BC748_1345 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
969894 |
24 |
2750 |
346.0 |
39.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.031 ± 0.053 | 0.808 ± 0.018 |
5.284 ± 0.034 | 6.432 ± 0.057 |
5.643 ± 0.041 | 6.12 ± 0.052 |
1.534 ± 0.021 | 8.31 ± 0.056 |
7.966 ± 0.091 | 8.966 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.105 ± 0.025 | 6.988 ± 0.056 |
3.3 ± 0.033 | 3.404 ± 0.025 |
2.978 ± 0.029 | 6.68 ± 0.048 |
6.253 ± 0.093 | 6.175 ± 0.033 |
0.994 ± 0.017 | 4.029 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
