
Sinocyclocheilus anshuiensis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii;
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
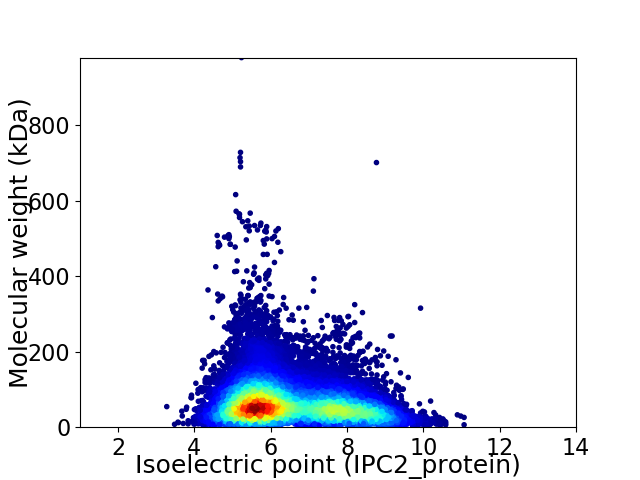
Virtual 2D-PAGE plot for 100275 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A671M871|A0A671M871_9TELE FXYD domain-containing ion transport regulator OS=Sinocyclocheilus anshuiensis OX=1608454 GN=LOC107665652 PE=3 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84PVEE5 pKa = 4.18GQCCGSCPSLPPIIIPGMDD24 pKa = 3.31SVEE27 pKa = 4.13LCTTQEE33 pKa = 4.28DD34 pKa = 4.73GCLPTDD40 pKa = 3.69GEE42 pKa = 4.15MGYY45 pKa = 9.99PEE47 pKa = 4.59AQAMDD52 pKa = 4.59EE53 pKa = 4.21YY54 pKa = 10.51PIQRR58 pKa = 11.84KK59 pKa = 8.62GDD61 pKa = 3.62RR62 pKa = 11.84LPFQIFPDD70 pKa = 3.82PVEE73 pKa = 4.32VVLSTEE79 pKa = 4.12GCLTCAPQEE88 pKa = 4.25KK89 pKa = 9.75EE90 pKa = 4.01RR91 pKa = 11.84LPSIVVEE98 pKa = 4.1PTDD101 pKa = 3.44VSEE104 pKa = 4.56VEE106 pKa = 4.27SGEE109 pKa = 4.3LRR111 pKa = 11.84WPPEE115 pKa = 4.09DD116 pKa = 3.39MDD118 pKa = 4.13FDD120 pKa = 4.24EE121 pKa = 7.53DD122 pKa = 3.63EE123 pKa = 5.21DD124 pKa = 5.44LFLEE128 pKa = 4.55QCIPPANIADD138 pKa = 3.45WGEE141 pKa = 4.12GEE143 pKa = 4.44EE144 pKa = 4.17EE145 pKa = 3.97SSIIEE150 pKa = 3.78NHH152 pKa = 5.08QQNTSLLDD160 pKa = 3.61LPRR163 pKa = 11.84SSGSSFEE170 pKa = 4.23YY171 pKa = 10.84
MM1 pKa = 7.64RR2 pKa = 11.84PVEE5 pKa = 4.18GQCCGSCPSLPPIIIPGMDD24 pKa = 3.31SVEE27 pKa = 4.13LCTTQEE33 pKa = 4.28DD34 pKa = 4.73GCLPTDD40 pKa = 3.69GEE42 pKa = 4.15MGYY45 pKa = 9.99PEE47 pKa = 4.59AQAMDD52 pKa = 4.59EE53 pKa = 4.21YY54 pKa = 10.51PIQRR58 pKa = 11.84KK59 pKa = 8.62GDD61 pKa = 3.62RR62 pKa = 11.84LPFQIFPDD70 pKa = 3.82PVEE73 pKa = 4.32VVLSTEE79 pKa = 4.12GCLTCAPQEE88 pKa = 4.25KK89 pKa = 9.75EE90 pKa = 4.01RR91 pKa = 11.84LPSIVVEE98 pKa = 4.1PTDD101 pKa = 3.44VSEE104 pKa = 4.56VEE106 pKa = 4.27SGEE109 pKa = 4.3LRR111 pKa = 11.84WPPEE115 pKa = 4.09DD116 pKa = 3.39MDD118 pKa = 4.13FDD120 pKa = 4.24EE121 pKa = 7.53DD122 pKa = 3.63EE123 pKa = 5.21DD124 pKa = 5.44LFLEE128 pKa = 4.55QCIPPANIADD138 pKa = 3.45WGEE141 pKa = 4.12GEE143 pKa = 4.44EE144 pKa = 4.17EE145 pKa = 3.97SSIIEE150 pKa = 3.78NHH152 pKa = 5.08QQNTSLLDD160 pKa = 3.61LPRR163 pKa = 11.84SSGSSFEE170 pKa = 4.23YY171 pKa = 10.84
Molecular weight: 18.94 kDa
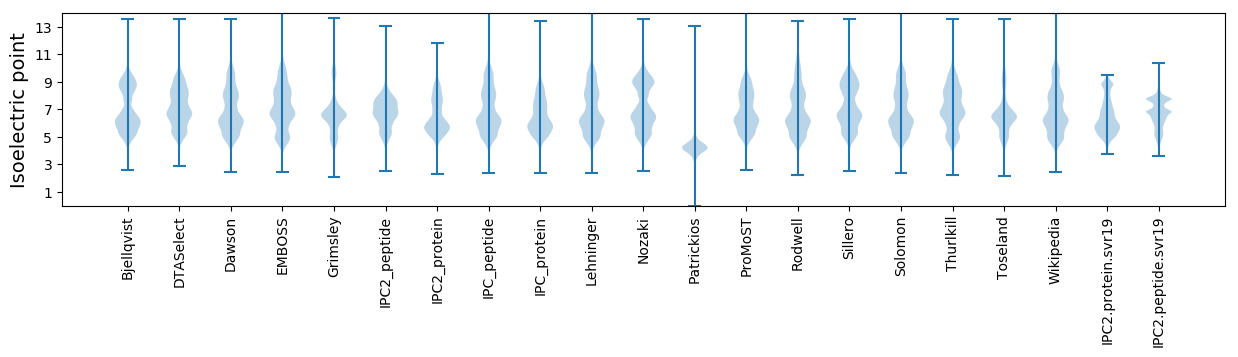
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A671QUH7|A0A671QUH7_9TELE Isoform of A0A671QUH6 Uncharacterized protein OS=Sinocyclocheilus anshuiensis OX=1608454 PE=4 SV=1
MM1 pKa = 7.45NMKK4 pKa = 10.17QNPTLHH10 pKa = 6.56IPPSKK15 pKa = 10.67GGSRR19 pKa = 11.84CPNKK23 pKa = 9.4PKK25 pKa = 10.12KK26 pKa = 9.78SRR28 pKa = 11.84RR29 pKa = 11.84VVEE32 pKa = 3.81GTGRR36 pKa = 11.84VGSHH40 pKa = 6.19GGAGRR45 pKa = 11.84SGSHH49 pKa = 6.22GGAGRR54 pKa = 11.84AGQPGGAGRR63 pKa = 11.84AGSHH67 pKa = 6.17GGAGRR72 pKa = 11.84AGSHH76 pKa = 6.17GGAGRR81 pKa = 11.84AGSHH85 pKa = 6.17GGAGRR90 pKa = 11.84AGSHH94 pKa = 6.17GGAGRR99 pKa = 11.84AGSHH103 pKa = 6.17GGAGRR108 pKa = 11.84AGSHH112 pKa = 6.17GGAGRR117 pKa = 11.84AGSHH121 pKa = 6.17GGAGRR126 pKa = 11.84AGSHH130 pKa = 6.17GGAGRR135 pKa = 11.84AGSHH139 pKa = 6.17GGAGRR144 pKa = 11.84AGSHH148 pKa = 6.17GGAGRR153 pKa = 11.84AGSHH157 pKa = 6.17GGAGRR162 pKa = 11.84AGSHH166 pKa = 6.17GGAGRR171 pKa = 11.84AGSHH175 pKa = 6.17GGAGRR180 pKa = 11.84AGSHH184 pKa = 6.17GGAGRR189 pKa = 11.84AGSHH193 pKa = 6.17GGAGRR198 pKa = 11.84AGSHH202 pKa = 6.17GGAGRR207 pKa = 11.84AGSHH211 pKa = 6.17GGAGRR216 pKa = 11.84AGSHH220 pKa = 6.17GGAGRR225 pKa = 11.84AGSHH229 pKa = 6.17GGAGRR234 pKa = 11.84AGSHH238 pKa = 6.17GGAGRR243 pKa = 11.84AGSHH247 pKa = 6.17GGAGRR252 pKa = 11.84AGSHH256 pKa = 6.17GGAGRR261 pKa = 11.84AGSHH265 pKa = 6.17GGAGRR270 pKa = 11.84AGSHH274 pKa = 6.17GGAGRR279 pKa = 11.84AGSHH283 pKa = 6.17GGAGRR288 pKa = 11.84AGSHH292 pKa = 6.17GGAGRR297 pKa = 11.84AGSHH301 pKa = 6.17GGAGRR306 pKa = 11.84AGSHH310 pKa = 6.17GGAGRR315 pKa = 11.84AGSHH319 pKa = 6.17GGAGRR324 pKa = 11.84AGSHH328 pKa = 6.17GGAGRR333 pKa = 11.84AGSHH337 pKa = 6.17GGAGRR342 pKa = 11.84AGSHH346 pKa = 6.17GGAGRR351 pKa = 11.84AGSHH355 pKa = 6.17GGAGRR360 pKa = 11.84AGSHH364 pKa = 6.17GGAGRR369 pKa = 11.84AGSHH373 pKa = 6.17GGAGRR378 pKa = 11.84AGRR381 pKa = 11.84RR382 pKa = 11.84GTGRR386 pKa = 11.84VGSHH390 pKa = 6.19GGAGRR395 pKa = 11.84SGKK398 pKa = 9.14IHH400 pKa = 6.37EE401 pKa = 5.24GKK403 pKa = 11.19GNMKK407 pKa = 8.96TT408 pKa = 3.4
MM1 pKa = 7.45NMKK4 pKa = 10.17QNPTLHH10 pKa = 6.56IPPSKK15 pKa = 10.67GGSRR19 pKa = 11.84CPNKK23 pKa = 9.4PKK25 pKa = 10.12KK26 pKa = 9.78SRR28 pKa = 11.84RR29 pKa = 11.84VVEE32 pKa = 3.81GTGRR36 pKa = 11.84VGSHH40 pKa = 6.19GGAGRR45 pKa = 11.84SGSHH49 pKa = 6.22GGAGRR54 pKa = 11.84AGQPGGAGRR63 pKa = 11.84AGSHH67 pKa = 6.17GGAGRR72 pKa = 11.84AGSHH76 pKa = 6.17GGAGRR81 pKa = 11.84AGSHH85 pKa = 6.17GGAGRR90 pKa = 11.84AGSHH94 pKa = 6.17GGAGRR99 pKa = 11.84AGSHH103 pKa = 6.17GGAGRR108 pKa = 11.84AGSHH112 pKa = 6.17GGAGRR117 pKa = 11.84AGSHH121 pKa = 6.17GGAGRR126 pKa = 11.84AGSHH130 pKa = 6.17GGAGRR135 pKa = 11.84AGSHH139 pKa = 6.17GGAGRR144 pKa = 11.84AGSHH148 pKa = 6.17GGAGRR153 pKa = 11.84AGSHH157 pKa = 6.17GGAGRR162 pKa = 11.84AGSHH166 pKa = 6.17GGAGRR171 pKa = 11.84AGSHH175 pKa = 6.17GGAGRR180 pKa = 11.84AGSHH184 pKa = 6.17GGAGRR189 pKa = 11.84AGSHH193 pKa = 6.17GGAGRR198 pKa = 11.84AGSHH202 pKa = 6.17GGAGRR207 pKa = 11.84AGSHH211 pKa = 6.17GGAGRR216 pKa = 11.84AGSHH220 pKa = 6.17GGAGRR225 pKa = 11.84AGSHH229 pKa = 6.17GGAGRR234 pKa = 11.84AGSHH238 pKa = 6.17GGAGRR243 pKa = 11.84AGSHH247 pKa = 6.17GGAGRR252 pKa = 11.84AGSHH256 pKa = 6.17GGAGRR261 pKa = 11.84AGSHH265 pKa = 6.17GGAGRR270 pKa = 11.84AGSHH274 pKa = 6.17GGAGRR279 pKa = 11.84AGSHH283 pKa = 6.17GGAGRR288 pKa = 11.84AGSHH292 pKa = 6.17GGAGRR297 pKa = 11.84AGSHH301 pKa = 6.17GGAGRR306 pKa = 11.84AGSHH310 pKa = 6.17GGAGRR315 pKa = 11.84AGSHH319 pKa = 6.17GGAGRR324 pKa = 11.84AGSHH328 pKa = 6.17GGAGRR333 pKa = 11.84AGSHH337 pKa = 6.17GGAGRR342 pKa = 11.84AGSHH346 pKa = 6.17GGAGRR351 pKa = 11.84AGSHH355 pKa = 6.17GGAGRR360 pKa = 11.84AGSHH364 pKa = 6.17GGAGRR369 pKa = 11.84AGSHH373 pKa = 6.17GGAGRR378 pKa = 11.84AGRR381 pKa = 11.84RR382 pKa = 11.84GTGRR386 pKa = 11.84VGSHH390 pKa = 6.19GGAGRR395 pKa = 11.84SGKK398 pKa = 9.14IHH400 pKa = 6.37EE401 pKa = 5.24GKK403 pKa = 11.19GNMKK407 pKa = 8.96TT408 pKa = 3.4
Molecular weight: 35.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
62118981 |
18 |
8680 |
619.5 |
69.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.313 ± 0.006 | 2.306 ± 0.006 |
5.261 ± 0.005 | 6.893 ± 0.008 |
3.865 ± 0.005 | 6.027 ± 0.009 |
2.671 ± 0.003 | 4.837 ± 0.005 |
5.898 ± 0.008 | 9.708 ± 0.009 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.485 ± 0.003 | 4.026 ± 0.004 |
5.268 ± 0.009 | 4.684 ± 0.006 |
5.438 ± 0.005 | 8.274 ± 0.009 |
5.526 ± 0.005 | 6.408 ± 0.006 |
1.17 ± 0.002 | 2.923 ± 0.004 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
