
Bos taurus papillomavirus 16
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyokappapapillomavirus; Dyokappapapillomavirus 3
Average proteome isoelectric point is 5.6
Get precalculated fractions of proteins
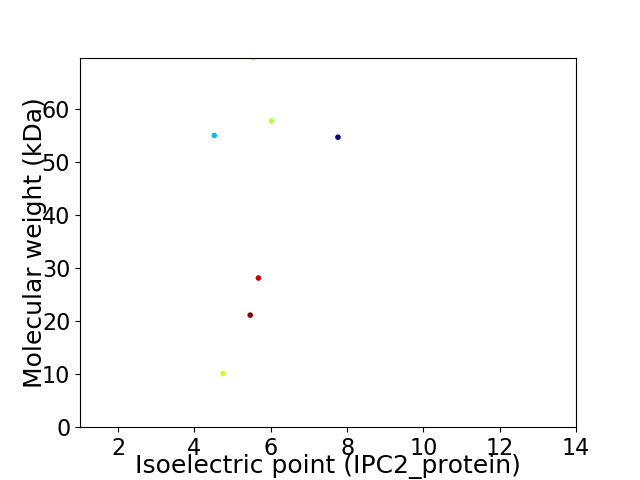
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2K201|A0A1B2K201_9PAPI Minor capsid protein L2 OS=Bos taurus papillomavirus 16 OX=1887214 GN=L2 PE=3 SV=1
MM1 pKa = 7.45HH2 pKa = 7.23GHH4 pKa = 6.49TPTLIDD10 pKa = 3.38IVLEE14 pKa = 3.98EE15 pKa = 4.95LPDD18 pKa = 4.29PVDD21 pKa = 3.95LQCQEE26 pKa = 3.79QLEE29 pKa = 4.36NEE31 pKa = 4.26EE32 pKa = 4.4EE33 pKa = 4.07EE34 pKa = 5.13GLLLDD39 pKa = 5.01DD40 pKa = 4.12YY41 pKa = 11.67VVYY44 pKa = 10.5ACCGHH49 pKa = 6.31CEE51 pKa = 3.89RR52 pKa = 11.84HH53 pKa = 5.28LKK55 pKa = 10.24VYY57 pKa = 10.39IRR59 pKa = 11.84GTVEE63 pKa = 3.75SVRR66 pKa = 11.84RR67 pKa = 11.84LQEE70 pKa = 3.95LLLSSLEE77 pKa = 4.36FVCPACGRR85 pKa = 11.84PRR87 pKa = 11.84RR88 pKa = 3.9
MM1 pKa = 7.45HH2 pKa = 7.23GHH4 pKa = 6.49TPTLIDD10 pKa = 3.38IVLEE14 pKa = 3.98EE15 pKa = 4.95LPDD18 pKa = 4.29PVDD21 pKa = 3.95LQCQEE26 pKa = 3.79QLEE29 pKa = 4.36NEE31 pKa = 4.26EE32 pKa = 4.4EE33 pKa = 4.07EE34 pKa = 5.13GLLLDD39 pKa = 5.01DD40 pKa = 4.12YY41 pKa = 11.67VVYY44 pKa = 10.5ACCGHH49 pKa = 6.31CEE51 pKa = 3.89RR52 pKa = 11.84HH53 pKa = 5.28LKK55 pKa = 10.24VYY57 pKa = 10.39IRR59 pKa = 11.84GTVEE63 pKa = 3.75SVRR66 pKa = 11.84RR67 pKa = 11.84LQEE70 pKa = 3.95LLLSSLEE77 pKa = 4.36FVCPACGRR85 pKa = 11.84PRR87 pKa = 11.84RR88 pKa = 3.9
Molecular weight: 10.12 kDa
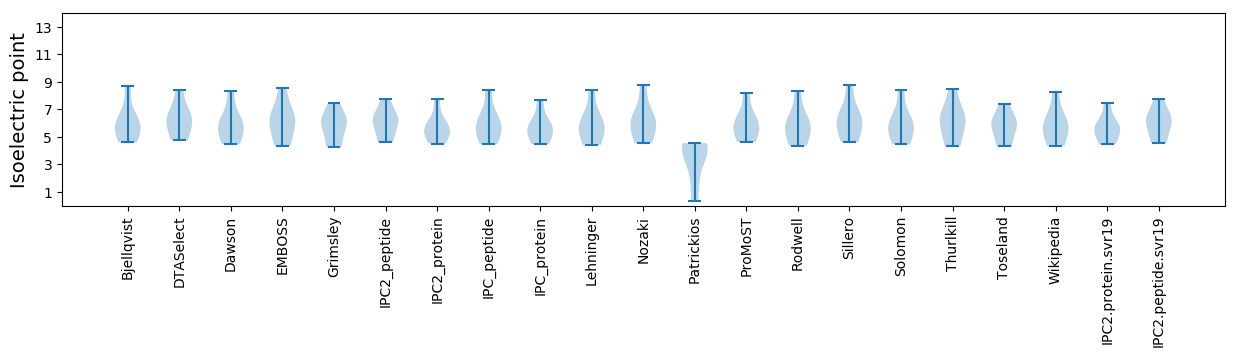
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2K253|A0A1B2K253_9PAPI Replication protein E1 OS=Bos taurus papillomavirus 16 OX=1887214 GN=E1 PE=3 SV=1
MM1 pKa = 6.91EE2 pKa = 4.72TLLARR7 pKa = 11.84LDD9 pKa = 3.83ALQEE13 pKa = 4.01QLLTLYY19 pKa = 10.56EE20 pKa = 4.43SDD22 pKa = 4.76SKK24 pKa = 11.61DD25 pKa = 2.85IDD27 pKa = 3.81DD28 pKa = 5.37QIKK31 pKa = 9.28HH32 pKa = 5.43WSLLRR37 pKa = 11.84QEE39 pKa = 5.1YY40 pKa = 10.16VLQHH44 pKa = 6.32FARR47 pKa = 11.84KK48 pKa = 9.53HH49 pKa = 5.3SISRR53 pKa = 11.84LGMQTVLPLQVSQHH67 pKa = 5.4KK68 pKa = 10.13AKK70 pKa = 10.42EE71 pKa = 4.16AIEE74 pKa = 3.81MVMYY78 pKa = 10.92LEE80 pKa = 4.29SLKK83 pKa = 11.07NSTFAVEE90 pKa = 4.35PWTLTDD96 pKa = 3.31TSRR99 pKa = 11.84EE100 pKa = 3.95MFIIPPKK107 pKa = 10.1NCFKK111 pKa = 11.04KK112 pKa = 10.75GGTTVDD118 pKa = 2.54VWYY121 pKa = 10.01DD122 pKa = 3.39CLQANSMHH130 pKa = 5.31YY131 pKa = 8.35TAWSYY136 pKa = 11.17IYY138 pKa = 10.91YY139 pKa = 8.26EE140 pKa = 4.64TEE142 pKa = 3.98SGWIKK147 pKa = 11.19AEE149 pKa = 3.92GQVDD153 pKa = 4.05YY154 pKa = 11.2NGCYY158 pKa = 10.07YY159 pKa = 10.78EE160 pKa = 5.33DD161 pKa = 4.28GNTRR165 pKa = 11.84IYY167 pKa = 8.98YY168 pKa = 9.28TRR170 pKa = 11.84FEE172 pKa = 5.33DD173 pKa = 4.41DD174 pKa = 3.11AQRR177 pKa = 11.84YY178 pKa = 7.04SKK180 pKa = 10.97NMEE183 pKa = 3.87WEE185 pKa = 4.17VHH187 pKa = 5.35YY188 pKa = 8.82KK189 pKa = 8.16THH191 pKa = 5.75VFSPVASVSSTSDD204 pKa = 3.01SWCGGEE210 pKa = 4.22PDD212 pKa = 3.79STGGPLSEE220 pKa = 4.19QHH222 pKa = 6.79SLPLDD227 pKa = 3.75NKK229 pKa = 8.39TSTWRR234 pKa = 11.84AAADD238 pKa = 3.49RR239 pKa = 11.84DD240 pKa = 4.2TNNNRR245 pKa = 11.84DD246 pKa = 3.27IRR248 pKa = 11.84NWSQDD253 pKa = 3.4TVDD256 pKa = 3.54GATVDD261 pKa = 2.92RR262 pKa = 11.84HH263 pKa = 4.57TVQRR267 pKa = 11.84ACGDD271 pKa = 3.5HH272 pKa = 5.75NRR274 pKa = 11.84RR275 pKa = 11.84GARR278 pKa = 11.84RR279 pKa = 11.84GRR281 pKa = 11.84GGLQGRR287 pKa = 11.84KK288 pKa = 6.29RR289 pKa = 11.84HH290 pKa = 5.95RR291 pKa = 11.84SRR293 pKa = 11.84EE294 pKa = 4.05EE295 pKa = 3.92GGSGQEE301 pKa = 4.59NRR303 pKa = 11.84QQSQTSKK310 pKa = 10.32RR311 pKa = 11.84PCEE314 pKa = 3.36RR315 pKa = 11.84RR316 pKa = 11.84LRR318 pKa = 11.84HH319 pKa = 5.19RR320 pKa = 11.84QRR322 pKa = 11.84PSGEE326 pKa = 4.09GQPQPRR332 pKa = 11.84PRR334 pKa = 11.84EE335 pKa = 4.07APAAAQQGGDD345 pKa = 3.31TSEE348 pKa = 4.18EE349 pKa = 4.16EE350 pKa = 3.7EE351 pKa = 5.81DD352 pKa = 4.56IRR354 pKa = 11.84QLPQRR359 pKa = 11.84GTPPHH364 pKa = 6.19SPEE367 pKa = 4.06RR368 pKa = 11.84PVDD371 pKa = 3.61ARR373 pKa = 11.84GHH375 pKa = 5.84GEE377 pKa = 3.96GAGEE381 pKa = 3.73PRR383 pKa = 11.84RR384 pKa = 11.84PRR386 pKa = 11.84RR387 pKa = 11.84AVRR390 pKa = 11.84PIEE393 pKa = 4.39HH394 pKa = 6.61NCDD397 pKa = 3.27TPVLLFRR404 pKa = 11.84GGANQLKK411 pKa = 8.84CARR414 pKa = 11.84YY415 pKa = 8.42RR416 pKa = 11.84WLRR419 pKa = 11.84QYY421 pKa = 11.65AHH423 pKa = 7.41LFLAISKK430 pKa = 7.5TWGWVGAVGLGASSRR445 pKa = 11.84LLLAFEE451 pKa = 4.74STTQRR456 pKa = 11.84NQFLDD461 pKa = 4.28TVPMPPGVTLGWGRR475 pKa = 11.84LSSLL479 pKa = 3.86
MM1 pKa = 6.91EE2 pKa = 4.72TLLARR7 pKa = 11.84LDD9 pKa = 3.83ALQEE13 pKa = 4.01QLLTLYY19 pKa = 10.56EE20 pKa = 4.43SDD22 pKa = 4.76SKK24 pKa = 11.61DD25 pKa = 2.85IDD27 pKa = 3.81DD28 pKa = 5.37QIKK31 pKa = 9.28HH32 pKa = 5.43WSLLRR37 pKa = 11.84QEE39 pKa = 5.1YY40 pKa = 10.16VLQHH44 pKa = 6.32FARR47 pKa = 11.84KK48 pKa = 9.53HH49 pKa = 5.3SISRR53 pKa = 11.84LGMQTVLPLQVSQHH67 pKa = 5.4KK68 pKa = 10.13AKK70 pKa = 10.42EE71 pKa = 4.16AIEE74 pKa = 3.81MVMYY78 pKa = 10.92LEE80 pKa = 4.29SLKK83 pKa = 11.07NSTFAVEE90 pKa = 4.35PWTLTDD96 pKa = 3.31TSRR99 pKa = 11.84EE100 pKa = 3.95MFIIPPKK107 pKa = 10.1NCFKK111 pKa = 11.04KK112 pKa = 10.75GGTTVDD118 pKa = 2.54VWYY121 pKa = 10.01DD122 pKa = 3.39CLQANSMHH130 pKa = 5.31YY131 pKa = 8.35TAWSYY136 pKa = 11.17IYY138 pKa = 10.91YY139 pKa = 8.26EE140 pKa = 4.64TEE142 pKa = 3.98SGWIKK147 pKa = 11.19AEE149 pKa = 3.92GQVDD153 pKa = 4.05YY154 pKa = 11.2NGCYY158 pKa = 10.07YY159 pKa = 10.78EE160 pKa = 5.33DD161 pKa = 4.28GNTRR165 pKa = 11.84IYY167 pKa = 8.98YY168 pKa = 9.28TRR170 pKa = 11.84FEE172 pKa = 5.33DD173 pKa = 4.41DD174 pKa = 3.11AQRR177 pKa = 11.84YY178 pKa = 7.04SKK180 pKa = 10.97NMEE183 pKa = 3.87WEE185 pKa = 4.17VHH187 pKa = 5.35YY188 pKa = 8.82KK189 pKa = 8.16THH191 pKa = 5.75VFSPVASVSSTSDD204 pKa = 3.01SWCGGEE210 pKa = 4.22PDD212 pKa = 3.79STGGPLSEE220 pKa = 4.19QHH222 pKa = 6.79SLPLDD227 pKa = 3.75NKK229 pKa = 8.39TSTWRR234 pKa = 11.84AAADD238 pKa = 3.49RR239 pKa = 11.84DD240 pKa = 4.2TNNNRR245 pKa = 11.84DD246 pKa = 3.27IRR248 pKa = 11.84NWSQDD253 pKa = 3.4TVDD256 pKa = 3.54GATVDD261 pKa = 2.92RR262 pKa = 11.84HH263 pKa = 4.57TVQRR267 pKa = 11.84ACGDD271 pKa = 3.5HH272 pKa = 5.75NRR274 pKa = 11.84RR275 pKa = 11.84GARR278 pKa = 11.84RR279 pKa = 11.84GRR281 pKa = 11.84GGLQGRR287 pKa = 11.84KK288 pKa = 6.29RR289 pKa = 11.84HH290 pKa = 5.95RR291 pKa = 11.84SRR293 pKa = 11.84EE294 pKa = 4.05EE295 pKa = 3.92GGSGQEE301 pKa = 4.59NRR303 pKa = 11.84QQSQTSKK310 pKa = 10.32RR311 pKa = 11.84PCEE314 pKa = 3.36RR315 pKa = 11.84RR316 pKa = 11.84LRR318 pKa = 11.84HH319 pKa = 5.19RR320 pKa = 11.84QRR322 pKa = 11.84PSGEE326 pKa = 4.09GQPQPRR332 pKa = 11.84PRR334 pKa = 11.84EE335 pKa = 4.07APAAAQQGGDD345 pKa = 3.31TSEE348 pKa = 4.18EE349 pKa = 4.16EE350 pKa = 3.7EE351 pKa = 5.81DD352 pKa = 4.56IRR354 pKa = 11.84QLPQRR359 pKa = 11.84GTPPHH364 pKa = 6.19SPEE367 pKa = 4.06RR368 pKa = 11.84PVDD371 pKa = 3.61ARR373 pKa = 11.84GHH375 pKa = 5.84GEE377 pKa = 3.96GAGEE381 pKa = 3.73PRR383 pKa = 11.84RR384 pKa = 11.84PRR386 pKa = 11.84RR387 pKa = 11.84AVRR390 pKa = 11.84PIEE393 pKa = 4.39HH394 pKa = 6.61NCDD397 pKa = 3.27TPVLLFRR404 pKa = 11.84GGANQLKK411 pKa = 8.84CARR414 pKa = 11.84YY415 pKa = 8.42RR416 pKa = 11.84WLRR419 pKa = 11.84QYY421 pKa = 11.65AHH423 pKa = 7.41LFLAISKK430 pKa = 7.5TWGWVGAVGLGASSRR445 pKa = 11.84LLLAFEE451 pKa = 4.74STTQRR456 pKa = 11.84NQFLDD461 pKa = 4.28TVPMPPGVTLGWGRR475 pKa = 11.84LSSLL479 pKa = 3.86
Molecular weight: 54.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2619 |
88 |
611 |
374.1 |
42.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.498 ± 0.54 | 2.482 ± 0.68 |
6.529 ± 0.464 | 6.758 ± 0.562 |
3.475 ± 0.523 | 6.682 ± 0.687 |
2.711 ± 0.307 | 4.582 ± 0.542 |
4.811 ± 0.834 | 7.827 ± 0.704 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.215 ± 0.411 | 4.124 ± 0.519 |
6.606 ± 1.089 | 4.506 ± 0.448 |
6.224 ± 0.856 | 7.102 ± 0.912 |
6.796 ± 0.41 | 5.613 ± 0.515 |
1.833 ± 0.339 | 3.627 ± 0.517 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
