
Wuhan Mosquito Virus 9
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
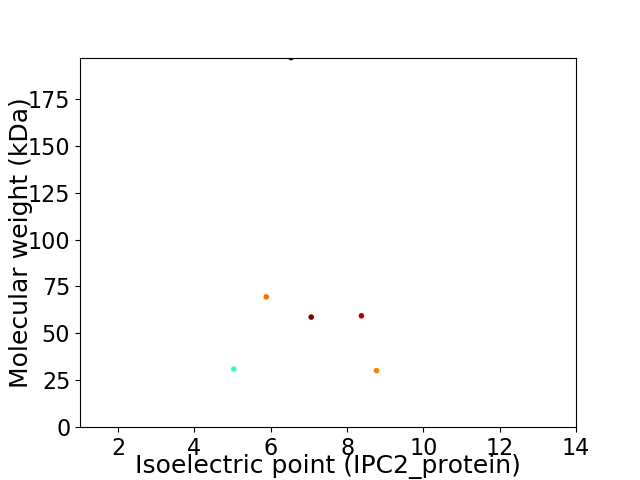
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KF56|A0A0B5KF56_9RHAB RNA-dependent RNA polymerase OS=Wuhan Mosquito Virus 9 OX=1608134 PE=4 SV=1
MM1 pKa = 7.44SKK3 pKa = 10.31ISICPVFVVPSYY15 pKa = 11.58NFMVTFSEE23 pKa = 4.52IIKK26 pKa = 10.43KK27 pKa = 9.94GAAISKK33 pKa = 9.87QILGRR38 pKa = 11.84IIYY41 pKa = 10.48SNVYY45 pKa = 9.02VDD47 pKa = 4.72KK48 pKa = 10.63EE49 pKa = 4.42SEE51 pKa = 4.28AVRR54 pKa = 11.84SVYY57 pKa = 9.15RR58 pKa = 11.84TGCLLSLSYY67 pKa = 10.56TGLSPVAWLVKK78 pKa = 9.53AAKK81 pKa = 9.43AKK83 pKa = 10.8SMSQLQLLKK92 pKa = 10.62KK93 pKa = 10.42LYY95 pKa = 8.68IPKK98 pKa = 9.19MEE100 pKa = 4.3SFITKK105 pKa = 9.99YY106 pKa = 10.41ISLSKK111 pKa = 10.54ISDD114 pKa = 3.74ATWVYY119 pKa = 10.8CRR121 pKa = 11.84LLSDD125 pKa = 3.62SALLKK130 pKa = 10.65LSASNEE136 pKa = 3.97PLATAVFVALCYY148 pKa = 10.3NPKK151 pKa = 10.39APDD154 pKa = 3.36MAVWEE159 pKa = 4.64IPSLQCISYY168 pKa = 8.76EE169 pKa = 4.11DD170 pKa = 3.69VVRR173 pKa = 11.84AHH175 pKa = 7.28CISEE179 pKa = 4.08AMMEE183 pKa = 4.29SPEE186 pKa = 4.24LLVEE190 pKa = 4.12QKK192 pKa = 10.62PLTKK196 pKa = 10.11EE197 pKa = 4.07AEE199 pKa = 4.12EE200 pKa = 4.17LTQKK204 pKa = 10.88LQTTAKK210 pKa = 9.82NCYY213 pKa = 8.74EE214 pKa = 3.93KK215 pKa = 10.64QQMVFRR221 pKa = 11.84TLFNQNRR228 pKa = 11.84SLLPKK233 pKa = 10.5NVDD236 pKa = 3.89SEE238 pKa = 4.19DD239 pKa = 3.56QEE241 pKa = 4.72QIEE244 pKa = 4.45QGDD247 pKa = 4.44GEE249 pKa = 5.01PGDD252 pKa = 5.92DD253 pKa = 4.88DD254 pKa = 4.79DD255 pKa = 6.75QPGQEE260 pKa = 3.91QDD262 pKa = 3.55AGGSTADD269 pKa = 4.01GSDD272 pKa = 3.35PAAGRR277 pKa = 11.84FEE279 pKa = 4.09
MM1 pKa = 7.44SKK3 pKa = 10.31ISICPVFVVPSYY15 pKa = 11.58NFMVTFSEE23 pKa = 4.52IIKK26 pKa = 10.43KK27 pKa = 9.94GAAISKK33 pKa = 9.87QILGRR38 pKa = 11.84IIYY41 pKa = 10.48SNVYY45 pKa = 9.02VDD47 pKa = 4.72KK48 pKa = 10.63EE49 pKa = 4.42SEE51 pKa = 4.28AVRR54 pKa = 11.84SVYY57 pKa = 9.15RR58 pKa = 11.84TGCLLSLSYY67 pKa = 10.56TGLSPVAWLVKK78 pKa = 9.53AAKK81 pKa = 9.43AKK83 pKa = 10.8SMSQLQLLKK92 pKa = 10.62KK93 pKa = 10.42LYY95 pKa = 8.68IPKK98 pKa = 9.19MEE100 pKa = 4.3SFITKK105 pKa = 9.99YY106 pKa = 10.41ISLSKK111 pKa = 10.54ISDD114 pKa = 3.74ATWVYY119 pKa = 10.8CRR121 pKa = 11.84LLSDD125 pKa = 3.62SALLKK130 pKa = 10.65LSASNEE136 pKa = 3.97PLATAVFVALCYY148 pKa = 10.3NPKK151 pKa = 10.39APDD154 pKa = 3.36MAVWEE159 pKa = 4.64IPSLQCISYY168 pKa = 8.76EE169 pKa = 4.11DD170 pKa = 3.69VVRR173 pKa = 11.84AHH175 pKa = 7.28CISEE179 pKa = 4.08AMMEE183 pKa = 4.29SPEE186 pKa = 4.24LLVEE190 pKa = 4.12QKK192 pKa = 10.62PLTKK196 pKa = 10.11EE197 pKa = 4.07AEE199 pKa = 4.12EE200 pKa = 4.17LTQKK204 pKa = 10.88LQTTAKK210 pKa = 9.82NCYY213 pKa = 8.74EE214 pKa = 3.93KK215 pKa = 10.64QQMVFRR221 pKa = 11.84TLFNQNRR228 pKa = 11.84SLLPKK233 pKa = 10.5NVDD236 pKa = 3.89SEE238 pKa = 4.19DD239 pKa = 3.56QEE241 pKa = 4.72QIEE244 pKa = 4.45QGDD247 pKa = 4.44GEE249 pKa = 5.01PGDD252 pKa = 5.92DD253 pKa = 4.88DD254 pKa = 4.79DD255 pKa = 6.75QPGQEE260 pKa = 3.91QDD262 pKa = 3.55AGGSTADD269 pKa = 4.01GSDD272 pKa = 3.35PAAGRR277 pKa = 11.84FEE279 pKa = 4.09
Molecular weight: 30.97 kDa
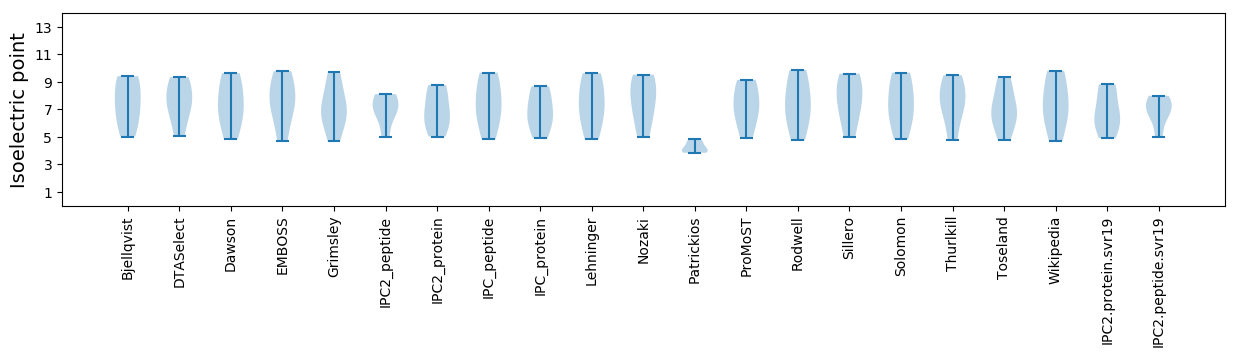
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KTR2|A0A0B5KTR2_9RHAB Glycoprotein OS=Wuhan Mosquito Virus 9 OX=1608134 GN=G PE=4 SV=1
MM1 pKa = 7.77EE2 pKa = 6.79DD3 pKa = 3.36IPMSMTLGEE12 pKa = 3.93ARR14 pKa = 11.84ARR16 pKa = 11.84RR17 pKa = 11.84AQVNVHH23 pKa = 5.97EE24 pKa = 5.13GEE26 pKa = 4.17GALIGGEE33 pKa = 4.02EE34 pKa = 4.49FARR37 pKa = 11.84MVGSISCTVTFAVEE51 pKa = 4.1LDD53 pKa = 3.71NRR55 pKa = 11.84SHH57 pKa = 6.31FLKK60 pKa = 10.87LISCLIFTEE69 pKa = 4.42SFYY72 pKa = 11.13ATSLSYY78 pKa = 10.89KK79 pKa = 10.17VIQTLATGLYY89 pKa = 8.71KK90 pKa = 10.75VLMDD94 pKa = 4.8PPYY97 pKa = 9.79ATPKK101 pKa = 10.73GSGGTQKK108 pKa = 10.04IHH110 pKa = 5.92VNYY113 pKa = 10.08KK114 pKa = 9.24FYY116 pKa = 10.97TRR118 pKa = 11.84TPCIRR123 pKa = 11.84PVDD126 pKa = 4.07LPKK129 pKa = 10.94VGGKK133 pKa = 10.2AIRR136 pKa = 11.84NFSVVGNAQRR146 pKa = 11.84CGFIEE151 pKa = 4.41YY152 pKa = 10.3NFSLTVHH159 pKa = 6.96HH160 pKa = 6.95LRR162 pKa = 11.84ANEE165 pKa = 3.74VGRR168 pKa = 11.84IEE170 pKa = 3.98GEE172 pKa = 4.02YY173 pKa = 8.76YY174 pKa = 10.93KK175 pKa = 10.76NVEE178 pKa = 4.11EE179 pKa = 4.67HH180 pKa = 7.23LSDD183 pKa = 4.66LKK185 pKa = 11.24CKK187 pKa = 9.65TLMKK191 pKa = 10.31RR192 pKa = 11.84IAEE195 pKa = 3.9IRR197 pKa = 11.84KK198 pKa = 9.67DD199 pKa = 3.45FTQFRR204 pKa = 11.84LADD207 pKa = 3.58YY208 pKa = 10.77LGLLNYY214 pKa = 8.22SHH216 pKa = 8.06KK217 pKa = 10.55GRR219 pKa = 11.84QLGKK223 pKa = 9.67IQSFPDD229 pKa = 3.14GWLEE233 pKa = 3.82SSLAGEE239 pKa = 4.59PSQPKK244 pKa = 10.12SPEE247 pKa = 3.74PLTRR251 pKa = 11.84GAKK254 pKa = 10.03AMAAIKK260 pKa = 9.4TRR262 pKa = 11.84VWKK265 pKa = 10.65RR266 pKa = 11.84KK267 pKa = 9.3GDD269 pKa = 3.46
MM1 pKa = 7.77EE2 pKa = 6.79DD3 pKa = 3.36IPMSMTLGEE12 pKa = 3.93ARR14 pKa = 11.84ARR16 pKa = 11.84RR17 pKa = 11.84AQVNVHH23 pKa = 5.97EE24 pKa = 5.13GEE26 pKa = 4.17GALIGGEE33 pKa = 4.02EE34 pKa = 4.49FARR37 pKa = 11.84MVGSISCTVTFAVEE51 pKa = 4.1LDD53 pKa = 3.71NRR55 pKa = 11.84SHH57 pKa = 6.31FLKK60 pKa = 10.87LISCLIFTEE69 pKa = 4.42SFYY72 pKa = 11.13ATSLSYY78 pKa = 10.89KK79 pKa = 10.17VIQTLATGLYY89 pKa = 8.71KK90 pKa = 10.75VLMDD94 pKa = 4.8PPYY97 pKa = 9.79ATPKK101 pKa = 10.73GSGGTQKK108 pKa = 10.04IHH110 pKa = 5.92VNYY113 pKa = 10.08KK114 pKa = 9.24FYY116 pKa = 10.97TRR118 pKa = 11.84TPCIRR123 pKa = 11.84PVDD126 pKa = 4.07LPKK129 pKa = 10.94VGGKK133 pKa = 10.2AIRR136 pKa = 11.84NFSVVGNAQRR146 pKa = 11.84CGFIEE151 pKa = 4.41YY152 pKa = 10.3NFSLTVHH159 pKa = 6.96HH160 pKa = 6.95LRR162 pKa = 11.84ANEE165 pKa = 3.74VGRR168 pKa = 11.84IEE170 pKa = 3.98GEE172 pKa = 4.02YY173 pKa = 8.76YY174 pKa = 10.93KK175 pKa = 10.76NVEE178 pKa = 4.11EE179 pKa = 4.67HH180 pKa = 7.23LSDD183 pKa = 4.66LKK185 pKa = 11.24CKK187 pKa = 9.65TLMKK191 pKa = 10.31RR192 pKa = 11.84IAEE195 pKa = 3.9IRR197 pKa = 11.84KK198 pKa = 9.67DD199 pKa = 3.45FTQFRR204 pKa = 11.84LADD207 pKa = 3.58YY208 pKa = 10.77LGLLNYY214 pKa = 8.22SHH216 pKa = 8.06KK217 pKa = 10.55GRR219 pKa = 11.84QLGKK223 pKa = 9.67IQSFPDD229 pKa = 3.14GWLEE233 pKa = 3.82SSLAGEE239 pKa = 4.59PSQPKK244 pKa = 10.12SPEE247 pKa = 3.74PLTRR251 pKa = 11.84GAKK254 pKa = 10.03AMAAIKK260 pKa = 9.4TRR262 pKa = 11.84VWKK265 pKa = 10.65RR266 pKa = 11.84KK267 pKa = 9.3GDD269 pKa = 3.46
Molecular weight: 30.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3936 |
269 |
1727 |
656.0 |
74.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.437 ± 0.569 | 2.109 ± 0.118 |
4.726 ± 0.342 | 6.072 ± 0.352 |
4.217 ± 0.286 | 5.31 ± 0.489 |
2.744 ± 0.425 | 6.021 ± 0.252 |
5.742 ± 0.447 | 10.722 ± 0.596 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.244 | 4.217 ± 0.351 |
5.081 ± 0.265 | 3.659 ± 0.386 |
5.285 ± 0.243 | 8.765 ± 0.457 |
5.691 ± 0.348 | 6.529 ± 0.448 |
1.804 ± 0.288 | 3.506 ± 0.366 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
