
Potato virus T (isolate Chenopodium amaranticolor/Peru/-/1992) (PVT)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Tepovirus; Potato virus T
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
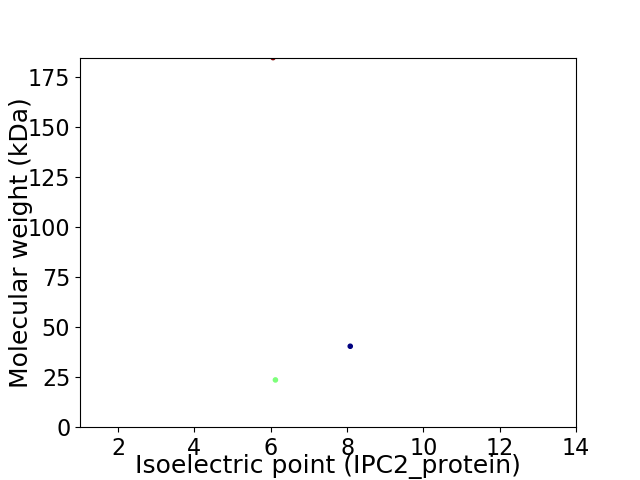
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
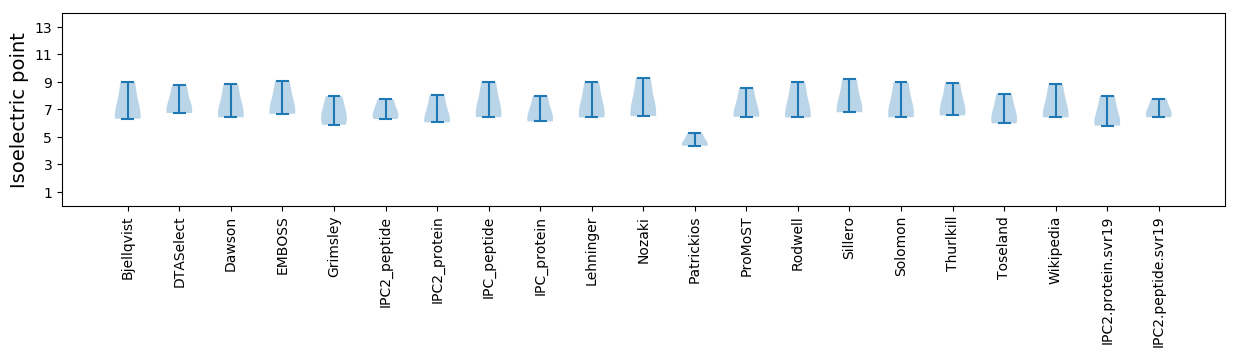
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B5ACE4|B5ACE4_PVT92 Coat protein OS=Potato virus T (isolate Chenopodium amaranticolor/Peru/-/1992) OX=1289386 PE=4 SV=1
MM1 pKa = 8.11DD2 pKa = 4.56PTTFVQIRR10 pKa = 11.84DD11 pKa = 3.57EE12 pKa = 4.23VLNLTVAAYY21 pKa = 10.15SSQWDD26 pKa = 3.96GQATQALKK34 pKa = 10.91DD35 pKa = 3.66GAKK38 pKa = 9.95EE39 pKa = 3.64QMLRR43 pKa = 11.84FLFGRR48 pKa = 11.84IAISSASRR56 pKa = 11.84NTIWPDD62 pKa = 3.19TEE64 pKa = 4.04IASEE68 pKa = 3.98DD69 pKa = 3.66LQIGMSAASAGPPPVAAAPISLIFRR94 pKa = 11.84VNFNSYY100 pKa = 9.59VKK102 pKa = 10.24MLIALSNTSTNSFVKK117 pKa = 10.64NKK119 pKa = 7.38TLRR122 pKa = 11.84QMCMPFAKK130 pKa = 10.12YY131 pKa = 10.18AYY133 pKa = 9.77GYY135 pKa = 9.57LSEE138 pKa = 4.53MGYY141 pKa = 8.36ATWAYY146 pKa = 9.99EE147 pKa = 4.66KK148 pKa = 10.22MPKK151 pKa = 9.97LCRR154 pKa = 11.84KK155 pKa = 9.06AKK157 pKa = 9.02WVAFDD162 pKa = 4.03FASGLLIDD170 pKa = 4.32TTMQLNDD177 pKa = 3.88DD178 pKa = 4.63EE179 pKa = 4.7KK180 pKa = 10.76TVIQGLGARR189 pKa = 11.84LFKK192 pKa = 9.87TQQSIQIADD201 pKa = 3.37STMDD205 pKa = 3.24GEE207 pKa = 4.83AINRR211 pKa = 11.84EE212 pKa = 3.94II213 pKa = 5.08
MM1 pKa = 8.11DD2 pKa = 4.56PTTFVQIRR10 pKa = 11.84DD11 pKa = 3.57EE12 pKa = 4.23VLNLTVAAYY21 pKa = 10.15SSQWDD26 pKa = 3.96GQATQALKK34 pKa = 10.91DD35 pKa = 3.66GAKK38 pKa = 9.95EE39 pKa = 3.64QMLRR43 pKa = 11.84FLFGRR48 pKa = 11.84IAISSASRR56 pKa = 11.84NTIWPDD62 pKa = 3.19TEE64 pKa = 4.04IASEE68 pKa = 3.98DD69 pKa = 3.66LQIGMSAASAGPPPVAAAPISLIFRR94 pKa = 11.84VNFNSYY100 pKa = 9.59VKK102 pKa = 10.24MLIALSNTSTNSFVKK117 pKa = 10.64NKK119 pKa = 7.38TLRR122 pKa = 11.84QMCMPFAKK130 pKa = 10.12YY131 pKa = 10.18AYY133 pKa = 9.77GYY135 pKa = 9.57LSEE138 pKa = 4.53MGYY141 pKa = 8.36ATWAYY146 pKa = 9.99EE147 pKa = 4.66KK148 pKa = 10.22MPKK151 pKa = 9.97LCRR154 pKa = 11.84KK155 pKa = 9.06AKK157 pKa = 9.02WVAFDD162 pKa = 4.03FASGLLIDD170 pKa = 4.32TTMQLNDD177 pKa = 3.88DD178 pKa = 4.63EE179 pKa = 4.7KK180 pKa = 10.76TVIQGLGARR189 pKa = 11.84LFKK192 pKa = 9.87TQQSIQIADD201 pKa = 3.37STMDD205 pKa = 3.24GEE207 pKa = 4.83AINRR211 pKa = 11.84EE212 pKa = 3.94II213 pKa = 5.08
Molecular weight: 23.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5ACE4|B5ACE4_PVT92 Coat protein OS=Potato virus T (isolate Chenopodium amaranticolor/Peru/-/1992) OX=1289386 PE=4 SV=1
MM1 pKa = 7.41EE2 pKa = 6.3LISVEE7 pKa = 3.93KK8 pKa = 10.46FRR10 pKa = 11.84RR11 pKa = 11.84QWEE14 pKa = 4.09EE15 pKa = 3.92RR16 pKa = 11.84EE17 pKa = 4.56SITGPVDD24 pKa = 2.97SGAIYY29 pKa = 10.29TNSAFHH35 pKa = 6.05NLKK38 pKa = 9.39TKK40 pKa = 7.48WHH42 pKa = 6.03VYY44 pKa = 10.09KK45 pKa = 10.59SEE47 pKa = 4.06CSIGLDD53 pKa = 3.85LPDD56 pKa = 3.73NGKK59 pKa = 10.11IISKK63 pKa = 9.9DD64 pKa = 2.93IPLFDD69 pKa = 4.49QEE71 pKa = 4.73EE72 pKa = 4.2IDD74 pKa = 6.11NIMKK78 pKa = 10.12DD79 pKa = 3.45DD80 pKa = 3.77KK81 pKa = 11.28QVFVHH86 pKa = 6.67LGAFVFGLVAHH97 pKa = 6.48FPVDD101 pKa = 3.7EE102 pKa = 4.2EE103 pKa = 4.85VEE105 pKa = 4.28GLVSIIDD112 pKa = 3.54KK113 pKa = 10.64RR114 pKa = 11.84RR115 pKa = 11.84TDD117 pKa = 3.39LRR119 pKa = 11.84RR120 pKa = 11.84ATLACRR126 pKa = 11.84KK127 pKa = 9.38IKK129 pKa = 10.17FVNGRR134 pKa = 11.84CAFMMKK140 pKa = 10.13PNFSVRR146 pKa = 11.84KK147 pKa = 8.93EE148 pKa = 3.88DD149 pKa = 4.97LRR151 pKa = 11.84DD152 pKa = 3.4GDD154 pKa = 4.32TFCAAIKK161 pKa = 10.15IKK163 pKa = 10.83NLGFEE168 pKa = 4.24GGFFPFSACGGVIYY182 pKa = 8.71RR183 pKa = 11.84TSNVSFAHH191 pKa = 6.6AVDD194 pKa = 3.33KK195 pKa = 10.78TFASRR200 pKa = 11.84TVHH203 pKa = 6.88DD204 pKa = 4.99LVGTDD209 pKa = 4.36ILSLDD214 pKa = 3.57QLDD217 pKa = 3.94RR218 pKa = 11.84ATLEE222 pKa = 3.99DD223 pKa = 3.88LEE225 pKa = 4.45EE226 pKa = 4.2VRR228 pKa = 11.84RR229 pKa = 11.84SPILRR234 pKa = 11.84LTAPDD239 pKa = 3.59EE240 pKa = 4.0RR241 pKa = 11.84VMIEE245 pKa = 3.76RR246 pKa = 11.84GNWFQKK252 pKa = 10.3KK253 pKa = 8.03PAIRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84SFGKK263 pKa = 10.06RR264 pKa = 11.84RR265 pKa = 11.84PQKK268 pKa = 9.68SSSVRR273 pKa = 11.84SASLPRR279 pKa = 11.84FSCSEE284 pKa = 3.56RR285 pKa = 11.84LEE287 pKa = 4.3RR288 pKa = 11.84GFEE291 pKa = 4.12SEE293 pKa = 4.27SLAGLVLDD301 pKa = 5.05QKK303 pKa = 11.12YY304 pKa = 10.49GPNNFCADD312 pKa = 3.29KK313 pKa = 10.65RR314 pKa = 11.84RR315 pKa = 11.84SPEE318 pKa = 3.72SDD320 pKa = 3.14SGSLQQSMGRR330 pKa = 11.84SSHH333 pKa = 6.28SGSEE337 pKa = 4.08GWGKK341 pKa = 8.5GTDD344 pKa = 3.34VAIPLRR350 pKa = 11.84KK351 pKa = 9.35NRR353 pKa = 11.84NIFSEE358 pKa = 4.3
MM1 pKa = 7.41EE2 pKa = 6.3LISVEE7 pKa = 3.93KK8 pKa = 10.46FRR10 pKa = 11.84RR11 pKa = 11.84QWEE14 pKa = 4.09EE15 pKa = 3.92RR16 pKa = 11.84EE17 pKa = 4.56SITGPVDD24 pKa = 2.97SGAIYY29 pKa = 10.29TNSAFHH35 pKa = 6.05NLKK38 pKa = 9.39TKK40 pKa = 7.48WHH42 pKa = 6.03VYY44 pKa = 10.09KK45 pKa = 10.59SEE47 pKa = 4.06CSIGLDD53 pKa = 3.85LPDD56 pKa = 3.73NGKK59 pKa = 10.11IISKK63 pKa = 9.9DD64 pKa = 2.93IPLFDD69 pKa = 4.49QEE71 pKa = 4.73EE72 pKa = 4.2IDD74 pKa = 6.11NIMKK78 pKa = 10.12DD79 pKa = 3.45DD80 pKa = 3.77KK81 pKa = 11.28QVFVHH86 pKa = 6.67LGAFVFGLVAHH97 pKa = 6.48FPVDD101 pKa = 3.7EE102 pKa = 4.2EE103 pKa = 4.85VEE105 pKa = 4.28GLVSIIDD112 pKa = 3.54KK113 pKa = 10.64RR114 pKa = 11.84RR115 pKa = 11.84TDD117 pKa = 3.39LRR119 pKa = 11.84RR120 pKa = 11.84ATLACRR126 pKa = 11.84KK127 pKa = 9.38IKK129 pKa = 10.17FVNGRR134 pKa = 11.84CAFMMKK140 pKa = 10.13PNFSVRR146 pKa = 11.84KK147 pKa = 8.93EE148 pKa = 3.88DD149 pKa = 4.97LRR151 pKa = 11.84DD152 pKa = 3.4GDD154 pKa = 4.32TFCAAIKK161 pKa = 10.15IKK163 pKa = 10.83NLGFEE168 pKa = 4.24GGFFPFSACGGVIYY182 pKa = 8.71RR183 pKa = 11.84TSNVSFAHH191 pKa = 6.6AVDD194 pKa = 3.33KK195 pKa = 10.78TFASRR200 pKa = 11.84TVHH203 pKa = 6.88DD204 pKa = 4.99LVGTDD209 pKa = 4.36ILSLDD214 pKa = 3.57QLDD217 pKa = 3.94RR218 pKa = 11.84ATLEE222 pKa = 3.99DD223 pKa = 3.88LEE225 pKa = 4.45EE226 pKa = 4.2VRR228 pKa = 11.84RR229 pKa = 11.84SPILRR234 pKa = 11.84LTAPDD239 pKa = 3.59EE240 pKa = 4.0RR241 pKa = 11.84VMIEE245 pKa = 3.76RR246 pKa = 11.84GNWFQKK252 pKa = 10.3KK253 pKa = 8.03PAIRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84SFGKK263 pKa = 10.06RR264 pKa = 11.84RR265 pKa = 11.84PQKK268 pKa = 9.68SSSVRR273 pKa = 11.84SASLPRR279 pKa = 11.84FSCSEE284 pKa = 3.56RR285 pKa = 11.84LEE287 pKa = 4.3RR288 pKa = 11.84GFEE291 pKa = 4.12SEE293 pKa = 4.27SLAGLVLDD301 pKa = 5.05QKK303 pKa = 11.12YY304 pKa = 10.49GPNNFCADD312 pKa = 3.29KK313 pKa = 10.65RR314 pKa = 11.84RR315 pKa = 11.84SPEE318 pKa = 3.72SDD320 pKa = 3.14SGSLQQSMGRR330 pKa = 11.84SSHH333 pKa = 6.28SGSEE337 pKa = 4.08GWGKK341 pKa = 8.5GTDD344 pKa = 3.34VAIPLRR350 pKa = 11.84KK351 pKa = 9.35NRR353 pKa = 11.84NIFSEE358 pKa = 4.3
Molecular weight: 40.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2177 |
213 |
1606 |
725.7 |
82.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.293 ± 1.386 | 1.791 ± 0.217 |
6.798 ± 0.296 | 7.304 ± 0.849 |
6.615 ± 0.528 | 5.374 ± 0.792 |
2.021 ± 0.537 | 5.926 ± 0.288 |
7.625 ± 0.723 | 9.371 ± 0.934 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.389 ± 0.608 | 3.95 ± 0.138 |
3.491 ± 0.206 | 2.848 ± 0.709 |
6.293 ± 1.233 | 7.671 ± 0.918 |
4.593 ± 0.674 | 5.926 ± 0.433 |
1.378 ± 0.15 | 2.343 ± 0.521 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
