
Acartia tonsa copepod circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 8.43
Get precalculated fractions of proteins
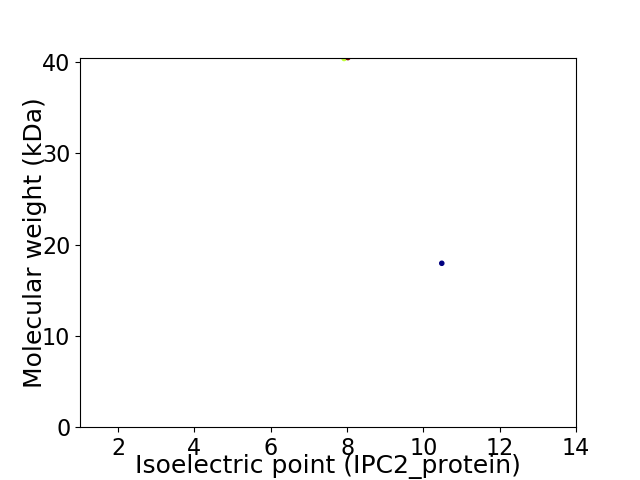
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L7P669|L7P669_9CIRC Uncharacterized protein OS=Acartia tonsa copepod circovirus OX=1168547 GN=AtCopCV_gp2 PE=4 SV=1
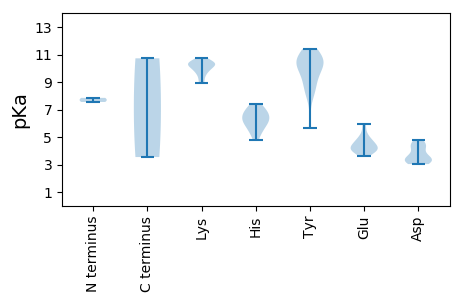
MM1 pKa = 7.55KK2 pKa = 10.02SLHH5 pKa = 5.86ISKK8 pKa = 9.92IEE10 pKa = 4.0FSQWPQSIKK19 pKa = 10.3PRR21 pKa = 11.84AKK23 pKa = 8.94RR24 pKa = 11.84AVGIFSLTKK33 pKa = 8.98MPSNATTNRR42 pKa = 11.84ARR44 pKa = 11.84GWCFTLNNPPPEE56 pKa = 4.67MISEE60 pKa = 4.09RR61 pKa = 11.84RR62 pKa = 11.84VVVPGHH68 pKa = 6.01VSGLDD73 pKa = 3.2AAIPTEE79 pKa = 4.28PNLPEE84 pKa = 5.13HH85 pKa = 6.38RR86 pKa = 11.84HH87 pKa = 4.79CDD89 pKa = 3.52YY90 pKa = 11.05RR91 pKa = 11.84VGQLEE96 pKa = 4.03RR97 pKa = 11.84GDD99 pKa = 4.0NGTLHH104 pKa = 5.65IQGYY108 pKa = 5.68YY109 pKa = 8.27HH110 pKa = 6.51HH111 pKa = 7.39RR112 pKa = 11.84VMKK115 pKa = 10.57SFNQMKK121 pKa = 10.32KK122 pKa = 10.43LIPNAHH128 pKa = 6.01WEE130 pKa = 4.1IAKK133 pKa = 10.33GSPKK137 pKa = 10.04QNRR140 pKa = 11.84DD141 pKa = 3.05YY142 pKa = 10.41CTKK145 pKa = 10.44EE146 pKa = 3.87DD147 pKa = 3.52TRR149 pKa = 11.84VPDD152 pKa = 3.4TRR154 pKa = 11.84PFEE157 pKa = 4.68EE158 pKa = 5.6GEE160 pKa = 4.41CPHH163 pKa = 6.26QGTRR167 pKa = 11.84FDD169 pKa = 4.79LRR171 pKa = 11.84QCADD175 pKa = 3.03VLANGGSLLDD185 pKa = 4.25LPGEE189 pKa = 4.18MVIRR193 pKa = 11.84YY194 pKa = 8.52HH195 pKa = 7.01RR196 pKa = 11.84GFSAYY201 pKa = 9.77RR202 pKa = 11.84SLFHH206 pKa = 7.24KK207 pKa = 10.28RR208 pKa = 11.84RR209 pKa = 11.84QEE211 pKa = 3.83APTVLWIYY219 pKa = 10.88GPTGSGKK226 pKa = 10.32SRR228 pKa = 11.84FASTLRR234 pKa = 11.84DD235 pKa = 3.14EE236 pKa = 5.96CYY238 pKa = 9.39WKK240 pKa = 10.35PPGKK244 pKa = 9.7WFDD247 pKa = 4.35GYY249 pKa = 10.87DD250 pKa = 3.52QEE252 pKa = 4.96PLVILDD258 pKa = 4.51DD259 pKa = 4.32YY260 pKa = 10.82RR261 pKa = 11.84SEE263 pKa = 3.8WAEE266 pKa = 3.6WGVFGQLLRR275 pKa = 11.84LLDD278 pKa = 4.61RR279 pKa = 11.84YY280 pKa = 10.45PLMVEE285 pKa = 4.04YY286 pKa = 10.56KK287 pKa = 9.53GGSVQFCSEE296 pKa = 5.04LIVITCDD303 pKa = 3.4RR304 pKa = 11.84PPSEE308 pKa = 4.64LWFSQGDD315 pKa = 3.24IGQLTRR321 pKa = 11.84RR322 pKa = 11.84ISHH325 pKa = 6.55VIHH328 pKa = 6.75AVGMWLPVEE337 pKa = 4.26PAVHH341 pKa = 6.15EE342 pKa = 4.64SIKK345 pKa = 10.78RR346 pKa = 11.84RR347 pKa = 11.84DD348 pKa = 3.68NIEE351 pKa = 3.54
MM1 pKa = 7.55KK2 pKa = 10.02SLHH5 pKa = 5.86ISKK8 pKa = 9.92IEE10 pKa = 4.0FSQWPQSIKK19 pKa = 10.3PRR21 pKa = 11.84AKK23 pKa = 8.94RR24 pKa = 11.84AVGIFSLTKK33 pKa = 8.98MPSNATTNRR42 pKa = 11.84ARR44 pKa = 11.84GWCFTLNNPPPEE56 pKa = 4.67MISEE60 pKa = 4.09RR61 pKa = 11.84RR62 pKa = 11.84VVVPGHH68 pKa = 6.01VSGLDD73 pKa = 3.2AAIPTEE79 pKa = 4.28PNLPEE84 pKa = 5.13HH85 pKa = 6.38RR86 pKa = 11.84HH87 pKa = 4.79CDD89 pKa = 3.52YY90 pKa = 11.05RR91 pKa = 11.84VGQLEE96 pKa = 4.03RR97 pKa = 11.84GDD99 pKa = 4.0NGTLHH104 pKa = 5.65IQGYY108 pKa = 5.68YY109 pKa = 8.27HH110 pKa = 6.51HH111 pKa = 7.39RR112 pKa = 11.84VMKK115 pKa = 10.57SFNQMKK121 pKa = 10.32KK122 pKa = 10.43LIPNAHH128 pKa = 6.01WEE130 pKa = 4.1IAKK133 pKa = 10.33GSPKK137 pKa = 10.04QNRR140 pKa = 11.84DD141 pKa = 3.05YY142 pKa = 10.41CTKK145 pKa = 10.44EE146 pKa = 3.87DD147 pKa = 3.52TRR149 pKa = 11.84VPDD152 pKa = 3.4TRR154 pKa = 11.84PFEE157 pKa = 4.68EE158 pKa = 5.6GEE160 pKa = 4.41CPHH163 pKa = 6.26QGTRR167 pKa = 11.84FDD169 pKa = 4.79LRR171 pKa = 11.84QCADD175 pKa = 3.03VLANGGSLLDD185 pKa = 4.25LPGEE189 pKa = 4.18MVIRR193 pKa = 11.84YY194 pKa = 8.52HH195 pKa = 7.01RR196 pKa = 11.84GFSAYY201 pKa = 9.77RR202 pKa = 11.84SLFHH206 pKa = 7.24KK207 pKa = 10.28RR208 pKa = 11.84RR209 pKa = 11.84QEE211 pKa = 3.83APTVLWIYY219 pKa = 10.88GPTGSGKK226 pKa = 10.32SRR228 pKa = 11.84FASTLRR234 pKa = 11.84DD235 pKa = 3.14EE236 pKa = 5.96CYY238 pKa = 9.39WKK240 pKa = 10.35PPGKK244 pKa = 9.7WFDD247 pKa = 4.35GYY249 pKa = 10.87DD250 pKa = 3.52QEE252 pKa = 4.96PLVILDD258 pKa = 4.51DD259 pKa = 4.32YY260 pKa = 10.82RR261 pKa = 11.84SEE263 pKa = 3.8WAEE266 pKa = 3.6WGVFGQLLRR275 pKa = 11.84LLDD278 pKa = 4.61RR279 pKa = 11.84YY280 pKa = 10.45PLMVEE285 pKa = 4.04YY286 pKa = 10.56KK287 pKa = 9.53GGSVQFCSEE296 pKa = 5.04LIVITCDD303 pKa = 3.4RR304 pKa = 11.84PPSEE308 pKa = 4.64LWFSQGDD315 pKa = 3.24IGQLTRR321 pKa = 11.84RR322 pKa = 11.84ISHH325 pKa = 6.55VIHH328 pKa = 6.75AVGMWLPVEE337 pKa = 4.26PAVHH341 pKa = 6.15EE342 pKa = 4.64SIKK345 pKa = 10.78RR346 pKa = 11.84RR347 pKa = 11.84DD348 pKa = 3.68NIEE351 pKa = 3.54
Molecular weight: 40.41 kDa
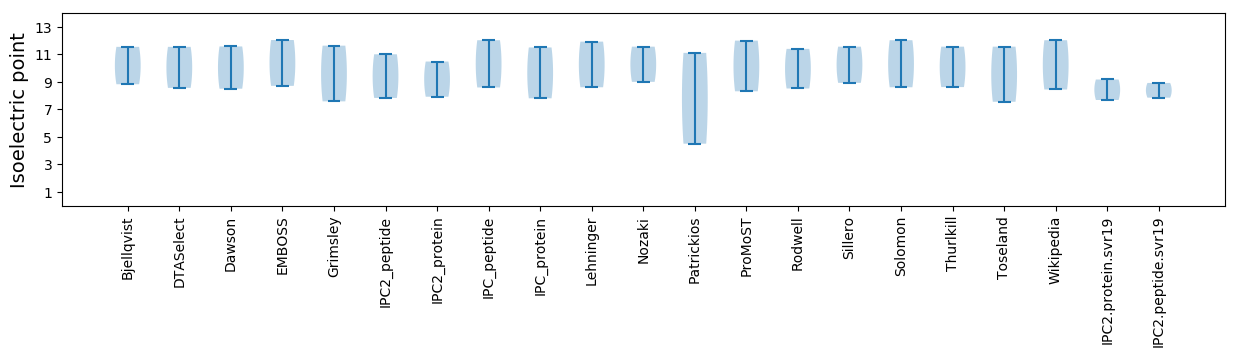
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L7P669|L7P669_9CIRC Uncharacterized protein OS=Acartia tonsa copepod circovirus OX=1168547 GN=AtCopCV_gp2 PE=4 SV=1
MM1 pKa = 7.85PNPHH5 pKa = 6.69FLEE8 pKa = 4.54PLNQAFNRR16 pKa = 11.84AAYY19 pKa = 10.03RR20 pKa = 11.84LLRR23 pKa = 11.84QAGGAATGAATTALYY38 pKa = 10.35EE39 pKa = 4.27YY40 pKa = 10.1ARR42 pKa = 11.84SRR44 pKa = 11.84ARR46 pKa = 11.84QMAYY50 pKa = 10.29GKK52 pKa = 10.18RR53 pKa = 11.84SYY55 pKa = 11.41SKK57 pKa = 10.61GRR59 pKa = 11.84TSYY62 pKa = 10.52GSRR65 pKa = 11.84YY66 pKa = 8.81RR67 pKa = 11.84PSYY70 pKa = 9.18RR71 pKa = 11.84KK72 pKa = 9.74KK73 pKa = 9.93RR74 pKa = 11.84YY75 pKa = 8.08SRR77 pKa = 11.84KK78 pKa = 9.33NRR80 pKa = 11.84SSMDD84 pKa = 3.19GASAVPPPSQSEE96 pKa = 4.32RR97 pKa = 11.84PRR99 pKa = 11.84QPSTTCWQSRR109 pKa = 11.84WCIRR113 pKa = 11.84HH114 pKa = 5.29GVMILIQVLVTWARR128 pKa = 11.84LSGLLPILSSALTPHH143 pKa = 5.39MHH145 pKa = 6.94KK146 pKa = 10.33SQAIAVRR153 pKa = 11.84ALLRR157 pKa = 11.84TKK159 pKa = 10.72
MM1 pKa = 7.85PNPHH5 pKa = 6.69FLEE8 pKa = 4.54PLNQAFNRR16 pKa = 11.84AAYY19 pKa = 10.03RR20 pKa = 11.84LLRR23 pKa = 11.84QAGGAATGAATTALYY38 pKa = 10.35EE39 pKa = 4.27YY40 pKa = 10.1ARR42 pKa = 11.84SRR44 pKa = 11.84ARR46 pKa = 11.84QMAYY50 pKa = 10.29GKK52 pKa = 10.18RR53 pKa = 11.84SYY55 pKa = 11.41SKK57 pKa = 10.61GRR59 pKa = 11.84TSYY62 pKa = 10.52GSRR65 pKa = 11.84YY66 pKa = 8.81RR67 pKa = 11.84PSYY70 pKa = 9.18RR71 pKa = 11.84KK72 pKa = 9.74KK73 pKa = 9.93RR74 pKa = 11.84YY75 pKa = 8.08SRR77 pKa = 11.84KK78 pKa = 9.33NRR80 pKa = 11.84SSMDD84 pKa = 3.19GASAVPPPSQSEE96 pKa = 4.32RR97 pKa = 11.84PRR99 pKa = 11.84QPSTTCWQSRR109 pKa = 11.84WCIRR113 pKa = 11.84HH114 pKa = 5.29GVMILIQVLVTWARR128 pKa = 11.84LSGLLPILSSALTPHH143 pKa = 5.39MHH145 pKa = 6.94KK146 pKa = 10.33SQAIAVRR153 pKa = 11.84ALLRR157 pKa = 11.84TKK159 pKa = 10.72
Molecular weight: 17.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
510 |
159 |
351 |
255.0 |
29.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.863 ± 2.704 | 1.961 ± 0.374 |
3.725 ± 1.646 | 5.098 ± 1.707 |
2.941 ± 0.895 | 7.059 ± 0.743 |
3.529 ± 0.539 | 4.706 ± 0.83 |
4.706 ± 0.161 | 8.039 ± 0.407 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.549 ± 0.317 | 2.941 ± 0.226 |
7.255 ± 0.179 | 4.314 ± 0.382 |
10.0 ± 1.705 | 8.039 ± 1.744 |
4.706 ± 0.507 | 4.902 ± 0.934 |
2.549 ± 0.352 | 4.118 ± 0.82 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
