
Gammapapillomavirus 14
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
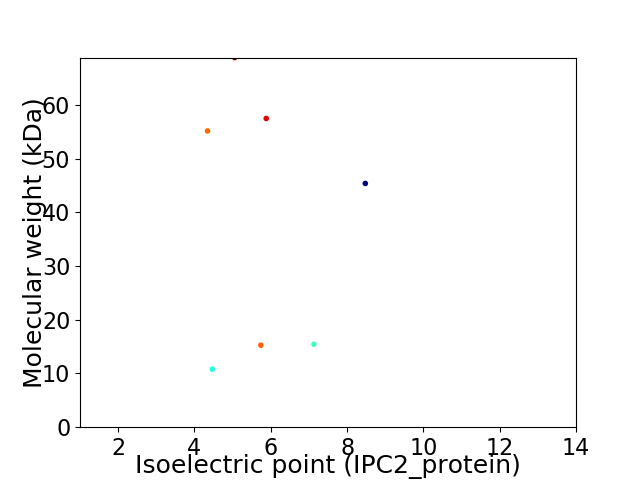
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALR3|A0A2D2ALR3_9PAPI E4 OS=Gammapapillomavirus 14 OX=1513259 GN=E4 PE=4 SV=1
MM1 pKa = 7.15SLKK4 pKa = 10.03RR5 pKa = 11.84KK6 pKa = 9.37KK7 pKa = 10.01RR8 pKa = 11.84ASPQDD13 pKa = 3.25LYY15 pKa = 11.2RR16 pKa = 11.84RR17 pKa = 11.84CAQGGDD23 pKa = 4.63CIPDD27 pKa = 3.39VQNKK31 pKa = 9.52YY32 pKa = 10.79EE33 pKa = 4.08NTTIADD39 pKa = 3.7WLLKK43 pKa = 10.38IFGSVIYY50 pKa = 10.31LGNLGIGTGRR60 pKa = 11.84GTGGSFGYY68 pKa = 10.34RR69 pKa = 11.84PFGSAGTGRR78 pKa = 11.84PAQEE82 pKa = 4.14LPIARR87 pKa = 11.84PNVVIDD93 pKa = 4.91PITPSSLIPVEE104 pKa = 4.41PGAPSIVPLVEE115 pKa = 3.8GTPDD119 pKa = 2.91IGFAGPDD126 pKa = 3.4AGPTVAGEE134 pKa = 4.4DD135 pKa = 3.35IEE137 pKa = 6.19LYY139 pKa = 10.41TLTTPTTDD147 pKa = 2.91VAGVGGGPSVITTEE161 pKa = 4.11EE162 pKa = 4.57LEE164 pKa = 4.16TAIIDD169 pKa = 3.69AHH171 pKa = 6.53PAPATPKK178 pKa = 9.75QVIYY182 pKa = 10.98DD183 pKa = 3.69SVAQVAIEE191 pKa = 3.86THH193 pKa = 6.12VNPFYY198 pKa = 11.45NPDD201 pKa = 3.56ANNLNIYY208 pKa = 9.84VDD210 pKa = 3.84PLITGDD216 pKa = 3.9TVGGSYY222 pKa = 10.75FEE224 pKa = 5.91DD225 pKa = 3.6IPLEE229 pKa = 4.12RR230 pKa = 11.84LDD232 pKa = 3.76LQSFEE237 pKa = 4.69VEE239 pKa = 4.27EE240 pKa = 5.01PPTEE244 pKa = 4.12STPTSIGGRR253 pKa = 11.84LTNRR257 pKa = 11.84ARR259 pKa = 11.84DD260 pKa = 3.54LYY262 pKa = 11.31SRR264 pKa = 11.84FIQQVPVNEE273 pKa = 4.45PDD275 pKa = 4.43FLVQPSRR282 pKa = 11.84LVQFEE287 pKa = 4.24IEE289 pKa = 4.63NPAFDD294 pKa = 5.29PDD296 pKa = 3.3VSLIFEE302 pKa = 4.89RR303 pKa = 11.84DD304 pKa = 3.43LEE306 pKa = 4.33GLRR309 pKa = 11.84AAPNEE314 pKa = 3.91EE315 pKa = 3.78FADD318 pKa = 3.76IVRR321 pKa = 11.84LSQPRR326 pKa = 11.84VTMTQEE332 pKa = 3.5GNVRR336 pKa = 11.84VSRR339 pKa = 11.84IGTKK343 pKa = 10.24AAITTRR349 pKa = 11.84SGLTVGPQVHH359 pKa = 5.99YY360 pKa = 10.75FMDD363 pKa = 4.37LSVIEE368 pKa = 4.75AVPEE372 pKa = 4.29TIEE375 pKa = 4.38LQTLNLPNDD384 pKa = 3.69SNTIVDD390 pKa = 4.86DD391 pKa = 4.44LLAHH395 pKa = 5.71TTFIDD400 pKa = 3.53PTNAATLHH408 pKa = 5.82YY409 pKa = 9.49EE410 pKa = 3.77EE411 pKa = 5.71SDD413 pKa = 3.33IYY415 pKa = 11.31DD416 pKa = 4.24ALPEE420 pKa = 4.49SFDD423 pKa = 3.38NTQLVLSTNEE433 pKa = 3.92EE434 pKa = 4.36GEE436 pKa = 4.53SLVFPAFTEE445 pKa = 4.36SGLKK449 pKa = 9.77TIFVPGFGSILKK461 pKa = 8.48TSPTIPPSSVIFSNDD476 pKa = 2.3TDD478 pKa = 3.7TFLLHH483 pKa = 6.34VADD486 pKa = 4.98INLDD490 pKa = 3.8YY491 pKa = 10.99DD492 pKa = 3.94LHH494 pKa = 6.43PALLPRR500 pKa = 11.84KK501 pKa = 9.31RR502 pKa = 11.84RR503 pKa = 11.84RR504 pKa = 11.84LDD506 pKa = 3.09AFF508 pKa = 3.85
MM1 pKa = 7.15SLKK4 pKa = 10.03RR5 pKa = 11.84KK6 pKa = 9.37KK7 pKa = 10.01RR8 pKa = 11.84ASPQDD13 pKa = 3.25LYY15 pKa = 11.2RR16 pKa = 11.84RR17 pKa = 11.84CAQGGDD23 pKa = 4.63CIPDD27 pKa = 3.39VQNKK31 pKa = 9.52YY32 pKa = 10.79EE33 pKa = 4.08NTTIADD39 pKa = 3.7WLLKK43 pKa = 10.38IFGSVIYY50 pKa = 10.31LGNLGIGTGRR60 pKa = 11.84GTGGSFGYY68 pKa = 10.34RR69 pKa = 11.84PFGSAGTGRR78 pKa = 11.84PAQEE82 pKa = 4.14LPIARR87 pKa = 11.84PNVVIDD93 pKa = 4.91PITPSSLIPVEE104 pKa = 4.41PGAPSIVPLVEE115 pKa = 3.8GTPDD119 pKa = 2.91IGFAGPDD126 pKa = 3.4AGPTVAGEE134 pKa = 4.4DD135 pKa = 3.35IEE137 pKa = 6.19LYY139 pKa = 10.41TLTTPTTDD147 pKa = 2.91VAGVGGGPSVITTEE161 pKa = 4.11EE162 pKa = 4.57LEE164 pKa = 4.16TAIIDD169 pKa = 3.69AHH171 pKa = 6.53PAPATPKK178 pKa = 9.75QVIYY182 pKa = 10.98DD183 pKa = 3.69SVAQVAIEE191 pKa = 3.86THH193 pKa = 6.12VNPFYY198 pKa = 11.45NPDD201 pKa = 3.56ANNLNIYY208 pKa = 9.84VDD210 pKa = 3.84PLITGDD216 pKa = 3.9TVGGSYY222 pKa = 10.75FEE224 pKa = 5.91DD225 pKa = 3.6IPLEE229 pKa = 4.12RR230 pKa = 11.84LDD232 pKa = 3.76LQSFEE237 pKa = 4.69VEE239 pKa = 4.27EE240 pKa = 5.01PPTEE244 pKa = 4.12STPTSIGGRR253 pKa = 11.84LTNRR257 pKa = 11.84ARR259 pKa = 11.84DD260 pKa = 3.54LYY262 pKa = 11.31SRR264 pKa = 11.84FIQQVPVNEE273 pKa = 4.45PDD275 pKa = 4.43FLVQPSRR282 pKa = 11.84LVQFEE287 pKa = 4.24IEE289 pKa = 4.63NPAFDD294 pKa = 5.29PDD296 pKa = 3.3VSLIFEE302 pKa = 4.89RR303 pKa = 11.84DD304 pKa = 3.43LEE306 pKa = 4.33GLRR309 pKa = 11.84AAPNEE314 pKa = 3.91EE315 pKa = 3.78FADD318 pKa = 3.76IVRR321 pKa = 11.84LSQPRR326 pKa = 11.84VTMTQEE332 pKa = 3.5GNVRR336 pKa = 11.84VSRR339 pKa = 11.84IGTKK343 pKa = 10.24AAITTRR349 pKa = 11.84SGLTVGPQVHH359 pKa = 5.99YY360 pKa = 10.75FMDD363 pKa = 4.37LSVIEE368 pKa = 4.75AVPEE372 pKa = 4.29TIEE375 pKa = 4.38LQTLNLPNDD384 pKa = 3.69SNTIVDD390 pKa = 4.86DD391 pKa = 4.44LLAHH395 pKa = 5.71TTFIDD400 pKa = 3.53PTNAATLHH408 pKa = 5.82YY409 pKa = 9.49EE410 pKa = 3.77EE411 pKa = 5.71SDD413 pKa = 3.33IYY415 pKa = 11.31DD416 pKa = 4.24ALPEE420 pKa = 4.49SFDD423 pKa = 3.38NTQLVLSTNEE433 pKa = 3.92EE434 pKa = 4.36GEE436 pKa = 4.53SLVFPAFTEE445 pKa = 4.36SGLKK449 pKa = 9.77TIFVPGFGSILKK461 pKa = 8.48TSPTIPPSSVIFSNDD476 pKa = 2.3TDD478 pKa = 3.7TFLLHH483 pKa = 6.34VADD486 pKa = 4.98INLDD490 pKa = 3.8YY491 pKa = 10.99DD492 pKa = 3.94LHH494 pKa = 6.43PALLPRR500 pKa = 11.84KK501 pKa = 9.31RR502 pKa = 11.84RR503 pKa = 11.84RR504 pKa = 11.84LDD506 pKa = 3.09AFF508 pKa = 3.85
Molecular weight: 55.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALT9|A0A2D2ALT9_9PAPI Protein E7 OS=Gammapapillomavirus 14 OX=1513259 GN=E7 PE=3 SV=1
MM1 pKa = 6.82QAEE4 pKa = 4.42TRR6 pKa = 11.84EE7 pKa = 4.26SLSARR12 pKa = 11.84FVAQQDD18 pKa = 3.44IQLSLIEE25 pKa = 4.76KK26 pKa = 9.56DD27 pKa = 3.53SKK29 pKa = 11.28NIEE32 pKa = 3.72DD33 pKa = 5.14HH34 pKa = 5.21IAYY37 pKa = 9.04WEE39 pKa = 4.62AIRR42 pKa = 11.84KK43 pKa = 8.73EE44 pKa = 4.13NVISYY49 pKa = 6.91YY50 pKa = 10.66ARR52 pKa = 11.84KK53 pKa = 8.91EE54 pKa = 3.72HH55 pKa = 6.29MGRR58 pKa = 11.84LGLQPLPTTAVSEE71 pKa = 4.35HH72 pKa = 6.4RR73 pKa = 11.84AKK75 pKa = 10.62EE76 pKa = 4.06AIKK79 pKa = 8.67MQIYY83 pKa = 10.01LRR85 pKa = 11.84SLARR89 pKa = 11.84SEE91 pKa = 4.2YY92 pKa = 10.36GRR94 pKa = 11.84EE95 pKa = 3.55PWTLTEE101 pKa = 4.84CSAEE105 pKa = 4.22MFNAPPRR112 pKa = 11.84DD113 pKa = 3.86CFKK116 pKa = 10.69KK117 pKa = 10.12KK118 pKa = 10.28PIMVTVWFDD127 pKa = 3.37NDD129 pKa = 3.41EE130 pKa = 4.56KK131 pKa = 11.63NSFPYY136 pKa = 8.96TAYY139 pKa = 10.29EE140 pKa = 3.86YY141 pKa = 10.15IYY143 pKa = 10.92YY144 pKa = 10.29KK145 pKa = 10.58DD146 pKa = 5.67DD147 pKa = 3.54NDD149 pKa = 2.78VWYY152 pKa = 10.03KK153 pKa = 10.02VQGLVDD159 pKa = 3.53HH160 pKa = 7.51DD161 pKa = 3.55GLYY164 pKa = 10.62FKK166 pKa = 10.49EE167 pKa = 4.28HH168 pKa = 6.38TGDD171 pKa = 3.12TVYY174 pKa = 11.18YY175 pKa = 10.5QLFQPDD181 pKa = 3.45ALKK184 pKa = 11.1YY185 pKa = 8.42GTTGQWTVKK194 pKa = 10.25FKK196 pKa = 11.09NQTVLASVTSSTRR209 pKa = 11.84SFSKK213 pKa = 10.06RR214 pKa = 11.84SEE216 pKa = 4.26VVSGGTTSHH225 pKa = 6.94ASTSKK230 pKa = 8.15EE231 pKa = 4.2TPRR234 pKa = 11.84GGPVEE239 pKa = 4.07TSEE242 pKa = 5.27DD243 pKa = 3.73PSSPSSTTPRR253 pKa = 11.84LRR255 pKa = 11.84SGRR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84QQGEE265 pKa = 3.98YY266 pKa = 10.18SSEE269 pKa = 3.92SRR271 pKa = 11.84AKK273 pKa = 10.06RR274 pKa = 11.84PRR276 pKa = 11.84VGSDD280 pKa = 2.95TAPTPDD286 pKa = 3.05EE287 pKa = 4.07VGRR290 pKa = 11.84RR291 pKa = 11.84SQSVAAHH298 pKa = 6.06GLPRR302 pKa = 11.84LRR304 pKa = 11.84RR305 pKa = 11.84LQEE308 pKa = 3.73EE309 pKa = 4.16ARR311 pKa = 11.84DD312 pKa = 3.78PPIILITGRR321 pKa = 11.84QNTLKK326 pKa = 10.39CWRR329 pKa = 11.84NRR331 pKa = 11.84CKK333 pKa = 10.65EE334 pKa = 3.82NEE336 pKa = 4.27KK337 pKa = 10.25ISKK340 pKa = 10.3LFMCCSSVWKK350 pKa = 9.72WLGTEE355 pKa = 4.31DD356 pKa = 4.47NGLSEE361 pKa = 4.51GKK363 pKa = 8.97MLIAFRR369 pKa = 11.84TIDD372 pKa = 3.44QRR374 pKa = 11.84TAFINSVTLPKK385 pKa = 10.46HH386 pKa = 5.41CTLSLGKK393 pKa = 10.06LDD395 pKa = 4.05SLL397 pKa = 4.89
MM1 pKa = 6.82QAEE4 pKa = 4.42TRR6 pKa = 11.84EE7 pKa = 4.26SLSARR12 pKa = 11.84FVAQQDD18 pKa = 3.44IQLSLIEE25 pKa = 4.76KK26 pKa = 9.56DD27 pKa = 3.53SKK29 pKa = 11.28NIEE32 pKa = 3.72DD33 pKa = 5.14HH34 pKa = 5.21IAYY37 pKa = 9.04WEE39 pKa = 4.62AIRR42 pKa = 11.84KK43 pKa = 8.73EE44 pKa = 4.13NVISYY49 pKa = 6.91YY50 pKa = 10.66ARR52 pKa = 11.84KK53 pKa = 8.91EE54 pKa = 3.72HH55 pKa = 6.29MGRR58 pKa = 11.84LGLQPLPTTAVSEE71 pKa = 4.35HH72 pKa = 6.4RR73 pKa = 11.84AKK75 pKa = 10.62EE76 pKa = 4.06AIKK79 pKa = 8.67MQIYY83 pKa = 10.01LRR85 pKa = 11.84SLARR89 pKa = 11.84SEE91 pKa = 4.2YY92 pKa = 10.36GRR94 pKa = 11.84EE95 pKa = 3.55PWTLTEE101 pKa = 4.84CSAEE105 pKa = 4.22MFNAPPRR112 pKa = 11.84DD113 pKa = 3.86CFKK116 pKa = 10.69KK117 pKa = 10.12KK118 pKa = 10.28PIMVTVWFDD127 pKa = 3.37NDD129 pKa = 3.41EE130 pKa = 4.56KK131 pKa = 11.63NSFPYY136 pKa = 8.96TAYY139 pKa = 10.29EE140 pKa = 3.86YY141 pKa = 10.15IYY143 pKa = 10.92YY144 pKa = 10.29KK145 pKa = 10.58DD146 pKa = 5.67DD147 pKa = 3.54NDD149 pKa = 2.78VWYY152 pKa = 10.03KK153 pKa = 10.02VQGLVDD159 pKa = 3.53HH160 pKa = 7.51DD161 pKa = 3.55GLYY164 pKa = 10.62FKK166 pKa = 10.49EE167 pKa = 4.28HH168 pKa = 6.38TGDD171 pKa = 3.12TVYY174 pKa = 11.18YY175 pKa = 10.5QLFQPDD181 pKa = 3.45ALKK184 pKa = 11.1YY185 pKa = 8.42GTTGQWTVKK194 pKa = 10.25FKK196 pKa = 11.09NQTVLASVTSSTRR209 pKa = 11.84SFSKK213 pKa = 10.06RR214 pKa = 11.84SEE216 pKa = 4.26VVSGGTTSHH225 pKa = 6.94ASTSKK230 pKa = 8.15EE231 pKa = 4.2TPRR234 pKa = 11.84GGPVEE239 pKa = 4.07TSEE242 pKa = 5.27DD243 pKa = 3.73PSSPSSTTPRR253 pKa = 11.84LRR255 pKa = 11.84SGRR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84QQGEE265 pKa = 3.98YY266 pKa = 10.18SSEE269 pKa = 3.92SRR271 pKa = 11.84AKK273 pKa = 10.06RR274 pKa = 11.84PRR276 pKa = 11.84VGSDD280 pKa = 2.95TAPTPDD286 pKa = 3.05EE287 pKa = 4.07VGRR290 pKa = 11.84RR291 pKa = 11.84SQSVAAHH298 pKa = 6.06GLPRR302 pKa = 11.84LRR304 pKa = 11.84RR305 pKa = 11.84LQEE308 pKa = 3.73EE309 pKa = 4.16ARR311 pKa = 11.84DD312 pKa = 3.78PPIILITGRR321 pKa = 11.84QNTLKK326 pKa = 10.39CWRR329 pKa = 11.84NRR331 pKa = 11.84CKK333 pKa = 10.65EE334 pKa = 3.82NEE336 pKa = 4.27KK337 pKa = 10.25ISKK340 pKa = 10.3LFMCCSSVWKK350 pKa = 9.72WLGTEE355 pKa = 4.31DD356 pKa = 4.47NGLSEE361 pKa = 4.51GKK363 pKa = 8.97MLIAFRR369 pKa = 11.84TIDD372 pKa = 3.44QRR374 pKa = 11.84TAFINSVTLPKK385 pKa = 10.46HH386 pKa = 5.41CTLSLGKK393 pKa = 10.06LDD395 pKa = 4.05SLL397 pKa = 4.89
Molecular weight: 45.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2378 |
94 |
601 |
339.7 |
38.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.056 ± 0.397 | 2.229 ± 0.73 |
6.56 ± 0.327 | 6.686 ± 0.676 |
4.71 ± 0.661 | 5.299 ± 0.674 |
1.766 ± 0.228 | 5.214 ± 0.672 |
5.341 ± 0.947 | 9.378 ± 0.628 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.598 ± 0.277 | 4.794 ± 0.612 |
6.476 ± 0.808 | 3.574 ± 0.418 |
5.509 ± 0.988 | 7.317 ± 0.525 |
6.728 ± 0.686 | 5.929 ± 0.46 |
1.262 ± 0.251 | 3.574 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
